Identifying individuals from biological mixtures to which they contributed is highly relevant in crime scene investigation and various biomedical research fields, but despite previous attempts, remains nearly impossible. Researchers at the Erasmus University Medical Center investigated the potential of using single-cell transcriptome sequencing (scRNA-seq), coupled with a dedicated bioinformatics pipeline (De-goulash), to solve this long-standing problem. The researchers developed a novel approach and tested it with scRNA-seq data that we de-novo generated from multi-person blood mixtures, and also in-silico mixtures they assembled from public single individual scRNA-seq datasets, involving different numbers, ratios, and bio-geographic ancestries of contributors. For all 2 up to 9-person balanced and imbalanced blood mixtures with ratios up to 1:60, the researchers achieved a clear single-cell separation according to the contributing individuals. For all separated mixture contributors, sex and bio-geographic ancestry (maternal, paternal, and bi-parental) were correctly determined. All separated contributors were correctly individually identified with court-acceptable statistical certainty using de-novo generated whole exome sequencing reference data. In this proof-of-concept study, these researchers demonstrate the feasibility of single-cell approaches to deconvolute biological mixtures and subsequently genetically characterise, and individually identify the separated mixture contributors. With further optimisation and implementation, this approach may eventually allow moving to challenging biological mixtures, including those found at crime scenes.
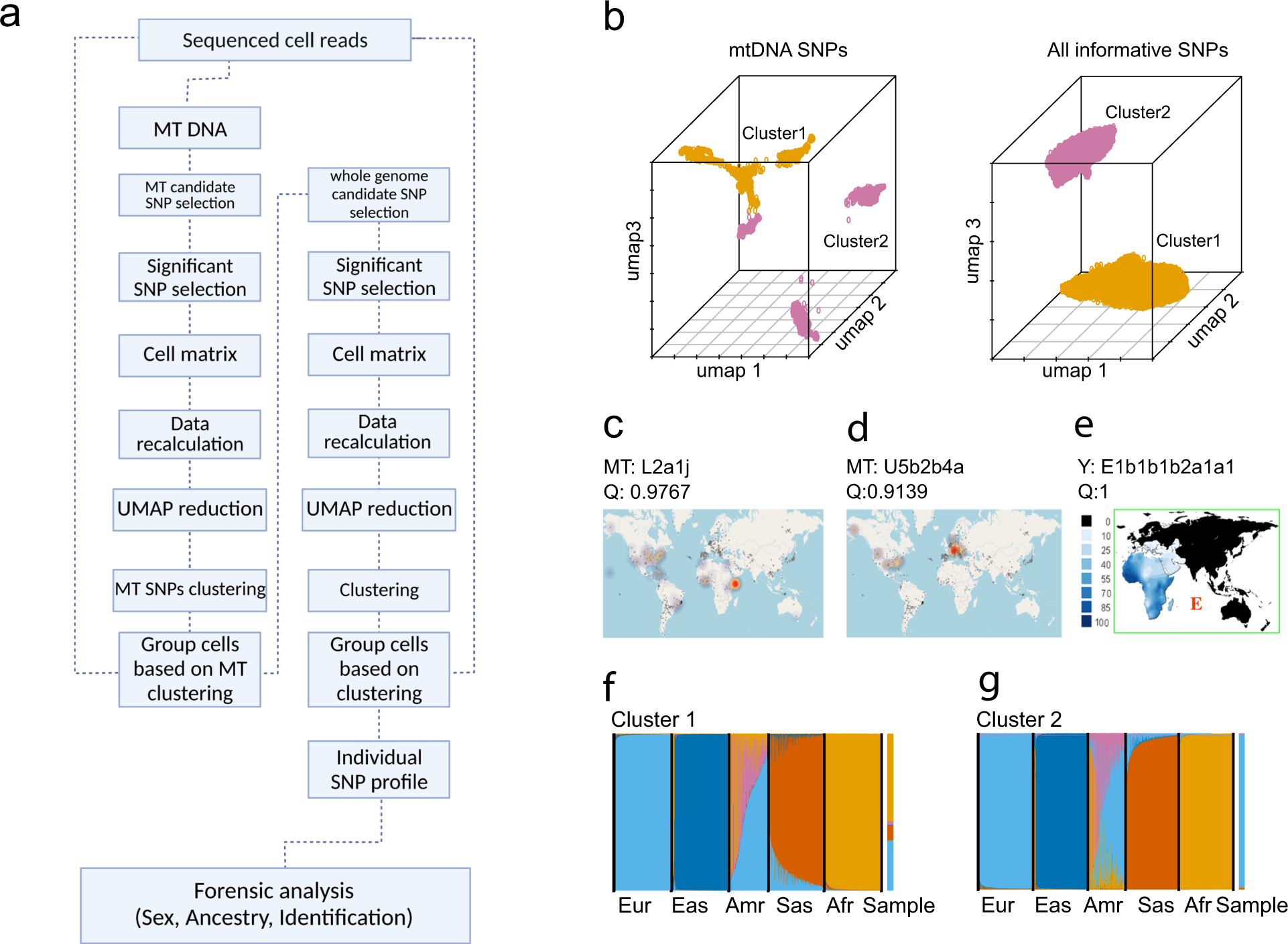
De-goulash bioinformatics pipeline for genetically deconvoluting a multi-person biological mixture with subsequent genetic characterization and individual identification of the separated mixture contributors based on single-cell transcriptome sequencing data. Pipeline description and application on balanced two-person blood mixture.
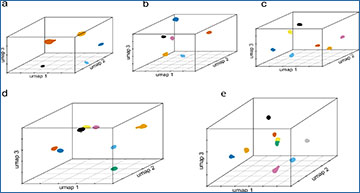
a The de-goulash pipeline workflow for single-cell-based mixture deconvolution with pre-processing of the scRNA-seq sequencing data in two iteration steps (mtDNA SNP-based separation followed by genome-wide SNP-based separation). b The 3D UMAP representation of the two-step single-cell separation process of a balanced two-person blood mixture (dataset M2) involving one male contributor of East African ancestry and the one female contributor of European ancestry. c EMPOP map of the worldwide distribution of mtDNA haplogroup L2a1j inferred from haplogroup-diagnostic mtDNA SNPs of cell cluster 1 with inferred African maternal ancestry. d EMPOP map of mtDNA haplogroup U5b2b4a inferred from haplogroup-diagnostic mtDNA SNPs of cluster 2 with inferred European maternal ancestry. e Literature map of Y haplogroup E inferred from haplogroup-diagnostic Y-SNPs of cell cluster 1 with inferred African paternal ancestry. Cluster 2 did not present a Y haplogroup due to female sex, as also revealed in the genetic sex analysis for cluster 2, while for cluster 1 male sex was obtained. f, g Biparental ancestry analysis with STRUCTURE of the genome-wide SNPs obtained per each of the cell clusters with continental reference population data (Eur: Europeans, Eas: East Asians, Amr: Native Americans, Afr: Sub-Saharan-Africans), the result for the cells clusters are denoted as Sample, result for cell cluster 1 demonstrates inferred admixed biparental ancestry with a major African ancestry, result for cell cluster 2 demonstrates European biparental ancestry. The maternal, paternal, and bi-parental genetic ancestries inferred from cell cluster 1 and 2 agree with the family-based ancestries of the two individuals involved in the mixture.
Availability – The bioinformatics pipeline de-goulash is available at: https://github.com/genid/de-goulash.
Kulhankova L, Montiel González D, Bindels E. et al. (2023) Single-cell transcriptome sequencing allows genetic separation, characterization and identification of individuals in multi-person biological mixtures. Commun Biol 6, 201 (2023). [article]