The systemic immune response to viral infection is shaped by master transcription factors, such as NF-κB, STAT1, or PU.1. Although long noncoding RNAs (lncRNAs) have been suggested as important regulators of transcription factor activity, their contributions to the systemic immunopathologies observed during SARS-CoV-2 infection have remained unknown. Researchers at Philipps University Marburg have now employed a targeted single-cell RNA sequencing approach to reveal lncRNAs differentially expressed in blood leukocytes during severe COVID-19. These results uncover the lncRNA PIRAT (PU.1-induced regulator of alarmin transcription) as a major PU.1 feedback-regulator in monocytes, governing the production of the alarmins S100A8/A9, key drivers of COVID-19 pathogenesis. Knockout and transgene expression, combined with chromatin-occupancy profiling, characterized PIRAT as a nuclear decoy RNA, keeping PU.1 from binding to alarmin promoters and promoting its binding to pseudogenes in naïve monocytes. NF-κB–dependent PIRAT down-regulation during COVID-19 consequently releases a transcriptional brake, fueling alarmin production. Alarmin expression is additionally enhanced by the up-regulation of the lncRNA LUCAT1, which promotes NF-κB–dependent gene expression at the expense of targets of the JAK-STAT pathway. These results suggest a major role of nuclear noncoding RNA networks in systemic antiviral responses to SARS-CoV-2 in humans.
Identification of human myeloid lineage-specific lincRNAs
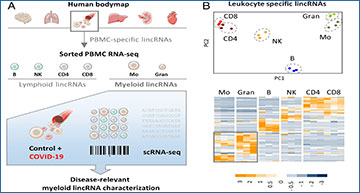
(A) Bulk and scRNA-seq strategy for the determination of myeloid lincRNAs relevant to COVID-19. (B) PCA (Upper) and hierarchical clustering (Lower, z-scores) of monocyte, granulocyte, B, NK, and CD4+ and CD8+ T cell lincRNA profiles. (C) qRT-PCR validation of PIRAT (LINC00211) and LUCAT1 as myeloid-specific lincRNAs (expression relative to human brain tissue). Horizontal bar indicates base-line (black) and twofold deviation from base-line (gray). (D) Relative abundance of PIRAT and LUCAT1 in RNA-seq profiles of hematopoietic stem cells (HSC), hematopoietic multipotent precursor cells (HMPC), common myeloid progenitors (CMP), granulocyte-macrophage progenitors (GMP), monocytes, and neutrophils. (E, Upper) PIRAT coexpression network, derived from RNA-seq data in B; (Lower) qRT-PCR analysis of PU.1 mRNA, PIRAT and LUCAT1 expression in PU.1 knockdown compared to control THP1 monocytes. (F and G) qRT-PCR analysis of lincRNA expression in response to indicated PAMPs and NF-κB inhibitor BAY-11-7082 (PAMP = 4 h LPS + polyI:C stimulation). (C–G) ≥3 independent experiments and one-way ANOVA test.
Aznaourova M, Schmerer N, Janga H, Zhang Z, Pauck K, Bushe J, Volkers SM, Wendisch D, Georg P, Ntini E, Aillaud M, Gündisch M, Mack E, Skevaki C, Keller C, Bauer C, Bertrams W, Marsico A, Nist A, Stiewe T, Gruber AD, Ruppert C, Li Y, Garn H, Sander LE, Schmeck B, Schulte LN. (2022) Single-cell RNA sequencing uncovers the nuclear decoy lincRNA PIRAT as a regulator of systemic monocyte immunity during COVID-19. Proc Natl Acad Sci U S A 119(36):e2120680119. [article]