Single-cell RNA sequencing (scRNA-seq) is currently one of the most powerful techniques available to study the transcriptional response of thousands of cells to an external perturbation. Researchers at the CSIC-Universitat de València performed a pseudotime analysis of SARS-CoV-2 infection using publicly available scRNA-seq data from human bronchial epithelial cells and colon and ileum organoids. These results reveal that, for most genes, the transcriptional response to SARS-CoV-2 infection follows a non-linear pattern characterized by an initial and a final down-regulatory phase separated by an intermediate up-regulatory stage. A correlation analysis of transcriptional profiles suggests a common mechanism regulating the mRNA levels of most genes. Interestingly, genes encoded in the mitochondria or involved in translation exhibited distinct pseudotime profiles. To explain their results, the researchers propose a simple model where nuclear export inhibition of nsp1-sensitive transcripts will be sufficient to explain the transcriptional shutdown of SARS-CoV-2 infected cells.
Proposed model to explain the transcriptional response of
infected cells to SARS-CoV-2 infection
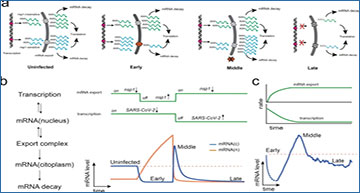
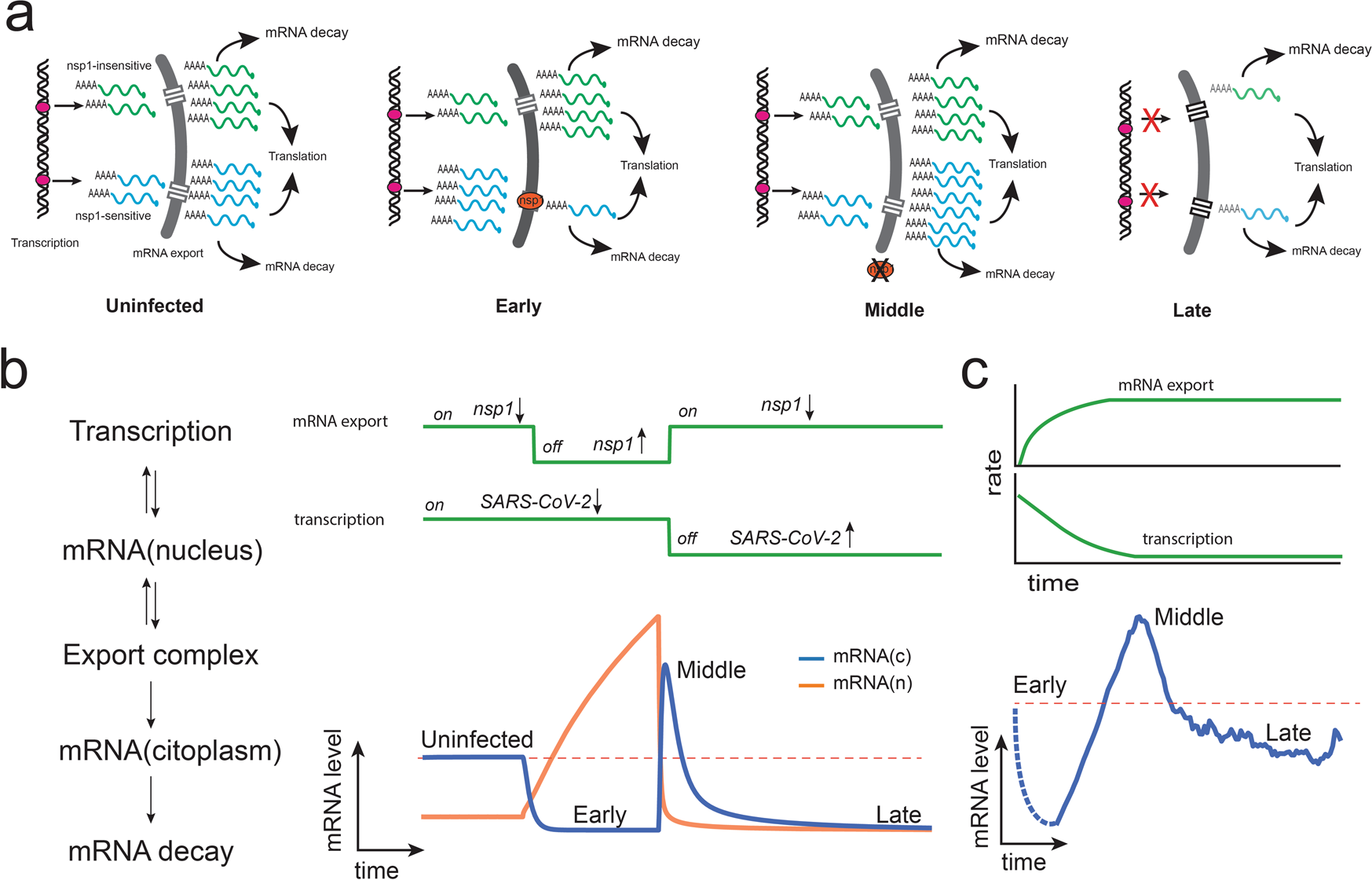
a Selective nuclear export of nsp1-sensitive transcripts will result in the degradation of cytoplasmic mRNAs and their accumulation in the nucleus. Once the blockage is removed, retained transcripts are released, resulting in a rapid increase in cytoplasmic mRNA levels. A shutdown of transcription during the late infection stage will result in a global decrease of host mRNAs. b A simplified dynamical model simulating nuclear mRNA export and transcription as on/off switches is enough to model the mean transcriptional profile. c Reinterpretation of the mean transcriptional profile of hBECs. In contrast to the on-off switch model presented in panel b, it is most likely that the shutdown of transcription and nuclear export occurs gradually, as shown in the upper panel. The dotted lines represent early infection states that could not be detected in the present study.
Gutiérrez PA, Elena SF. (2022) Single-cell RNA-sequencing data analysis reveals a highly correlated triphasic transcriptional response to SARS-CoV-2 infection. Commun Biol 5(1):1302.[article]