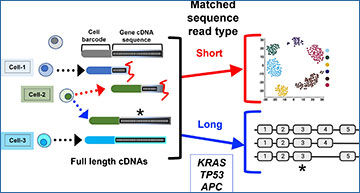
In this proof-of-concept study, researchers from the Stanford University School of Medicine have developed a single-cell method that provides genotypes of somatic alterations found in coding regions of messenger RNAs and integrates these transcript-based variants with their matching cell transcriptomes. The researchers used nanopore adaptive sampling on single-cell complementary DNA libraries to validate coding variants in target gene transcripts, and short-read sequencing to characterize cell types harboring the mutations. CRISPR edits for 16 targets were identified using a cancer cell line, and known variants in the cell line were validated using a 352-gene panel. Variants in primary cancer samples were validated using target gene panels ranging from 161 to 529 genes. A gene rearrangement was also identified in one patient, with the rearrangement occurring in two distinct tumor sites.
Availability – Scripts for analysis are publicly available on GitHub (https://github.com/sgtc-stanford/scCRISPR) (24) and Zenodo (https://zenodo.org/badge/latestdoi/365008149)
Grimes SM, Kim HS, Roy S, Sathe A, Ayala CI, Bai X, Almeda-Notestine AF, Haebe S, Shree T, Levy R, Lau BT, Ji HP. (2023) Single-cell multi-gene identification of somatic mutations and gene rearrangements in cancer NAR Cancer 5(3), zcad034. [article]