Cortical development in humans is a meticulously regulated process that unfolds over months of prenatal and years of postnatal life. This process encompasses the genesis, differentiation, and maturation of various cellular lineages that are critical to the complex structure and function of the brain and are affected in neurodevelopmental conditions. An area of active investigation is understanding the molecular mechanisms that define the developmental trajectory of specific cortical lineages. Recently, research in this field has extensively leveraged single-cell genomics, but its application has been largely confined to studying the second trimester of human cortical development.
Recent advances in technology, such as single-nucleus RNA sequencing (snRNA-seq), create new possibilities to explore previously uncharted territories of cortical development. These innovative tools can capture the molecular progression of cortical lineages across the life span, creating a more comprehensive and precise transcriptomic atlas of human cortical development. Furthermore, the integration of snRNA-seq and single-nucleus chromatin accessibility data allows for the identification of gene regulatory networks and transcription factors that are instrumental in shaping specific cortical lineages. This comprehensive approach could potentially unveil sex- and region-specific features and presents an unprecedented opportunity to shed light on lineage-specific susceptibility to neurodevelopmental and psychiatric conditions, such as autism spectrum disorder (ASD).
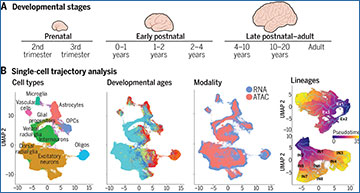
University of California, San Francisco researchers generated snRNA-seq data from human cortical samples during prenatal and postnatal stages of development and integrated these data with previously published datasets. Their study involved the analysis of >700,000 snRNA-seq profiles sourced from 169 tissue samples and 106 donors. Using single-cell trajectory analysis, the researchers identified developmental programs linked to the genesis of specific cortical cell types, including subtypes of excitatory neurons, interneurons, glial cells, and brain vasculature. They also determined sex- and region-specific developmental transcriptomic programs used by specific cortical lineages. By intersecting lineage-specific transcriptomic profiles with single-nucleus chromatin accessibility data, they defined enhancer gene regulatory networks and transcription factors pivotal to the commitment of defined cortical lineages. Using their insight into lineage-specific molecular developmental programs, they identified cell lineages and developmental stages implicated in the risk of neurodevelopmental disorders and observed that lineage-specific gene expression programs up-regulated in female cells are particularly enriched for the genetic risk factors of autism. This finding suggests that males may have an increased susceptibility to haploinsufficiency in ASD, which could provide a plausible explanation for the observed increased incidence of autism in males.
Single-cell genomics analysis of human cortical development across prenatal and postnatal life

(A) Developmental stages captured in this study. (B) Identification of cell types and lineages across developmental stages and molecular modalities. UMAP, uniform manifold approximation and projection; OPCs, oligodendrocyte precursor cells; Oligos, oligodendrocytes; ATAC, assay for transposase-accessible chromatin. (C) Discovery of lineage-specific developmental genes and enhancer gene regulatory networks. L2-3, upper-layer intratelencephalic projection neurons; PV-BSK, parvalbumin basket interneurons.
This study illuminates the molecular changes underlying the development human cortical lineages. By integrating single-nucleus RNA expression and chromatin accessibility profiling, these researchers charted a comprehensive transcriptomic atlas of cortical lineages across prenatal and postnatal development, identified key transcriptional networks, highlighted sex-specific developmental changes, and defined cell types and developmental stages most enriched for genetic risk factors of neurodevelopmental diseases. Their results shed light on lineage-specific mechanisms of normal cortical development, the genetic vulnerabilities to developmental brain disorders, and the role of sexually dimorphic gene expression in the pathogenesis of autism.
Velmeshev D, Perez Y, Yan Z, Valencia JE, Castaneda-Castellanos DR, Wang L, Schirmer L, Mayer S, Wick B, Wang S, Nowakowski TJ, Paredes M, Huang EJ, Kriegstein AR. (2023) Single-cell analysis of prenatal and postnatal human cortical development. Science 382(6667):eadf0834. [abstract]