Unsupervised clustering of single-cell RNA-sequencing data enables the identification and discovery of distinct cell populations. However, the most widely used clustering algorithms are heuristic and do not formally account for statistical uncertainty. Many popular pipelines use clustering stability methods to assess the algorithms’ output and decide on the number of clusters. However, we find that by not addressing known sources of variability in a statistically rigorous manner, these analyses lead to overconfidence in the discovery of novel cell-types.
Researchers from Harvard University and the Dana Farber Cancer Institute extend a previous method for Gaussian data, Significance of Hierarchical Clustering (SHC), to propose a model-based hypothesis testing approach that incorporates significance analysis into the clustering algorithm and permits statistical evaluation of clusters as distinct cell populations. The researchers also adapt this approach to permit statistical assessment on the clusters reported by any algorithm. They benchmarked their approach on real-world datasets against popular clustering workflows, demonstrating improved performance. To show its practical utility, the researchers applied it to the Human Lung Cell Atlas and an atlas of the mouse cerebellar cortex. They identified several cases of over-clustering, leading to false discoveries, as well as under-clustering, resulting in the failure to identify new subpopulations that their method was able to detect.
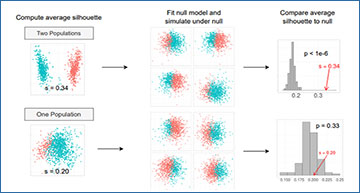
Schematic illustrating this approach to significance analysis for clustering
(A) Schematic of the test used by our approach to decide whether a proposed two-way split is significant. We show two examples, one in which two distinct populations were simulated (top) and one in which only one population was simulated (bottom). The schematics show how we use hierarchical clustering to divide the data into two, fit a single parametric model to the data, simulate 100 datasets under that model, and cluster each simulated dataset and compute the average silhouette. We then compare the average silhouette of the observed clusters to this empirical null distribution to decide whether to reject the null hypothesis. (B) Schematic of sc-SHC. We hierarchically cluster all the cells, and carry out significance analysis to decide whether to split the root node into the two clusters denoted by blue and red. We stop if we fail to reject the null hypothesis, and otherwise recursively continue performing tests to decide if we split each node or not.
Grabski IN, Street K, Irizarry RA. (2022) Significance Analysis for Clustering with Single-Cell RNA-Sequencing Data. bioRXiv [online preprint]. [abstract]