Single-cell technologies have revolutionised biological research and applications. As they continue to evolve with multi-omics and spatial resolution, analysing single-cell datasets is becoming increasingly complex. For biologists lacking expert data analysis resources, the problem is even more crucial, even for the simplest single-cell transcriptomics datasets.
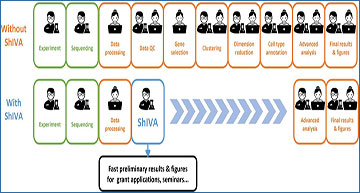
Researchers at INSERM, CIML have developed ShIVA, an interface for the analysis of single-cell RNA-seq and CITE-seq data specifically dedicated to biologists. Intuitive, iterative and documented by video tutorials, ShIVA allows biologists to follow a robust and reproducible analysis process, mostly based on the Seurat v4 R package, to fully explore and quantify their dataset, to produce useful figures and tables and to export their work to allow more complex analyses performed by experts.
ShIVA reduces single-cell genomics data analysis time
by placing the biologist project leader in power
ShIVA enables biologists to perform state-of-the-art exploratory analyses of their single-cell genomics datasets in a user-friendly interface.
Availability -Code and documentation are available from the GitHub repository: https://github.com/CIML-bioinformatic/ShIVA.
Aussel R, Asif M, Chenag S, Jaeger S, Milpied P, Spinelli L. (2022) ShIVA – A user-friendly and interactive interface giving biologists control over their single-cell RNA-seq data. bioRXiv [online preprint]. [article]