The advance in single-cell RNA sequencing technology has enhanced the analysis of cell development by profiling heterogeneous cells in individual cell resolution. In recent years, many trajectory inference methods have been developed. They have focused on using the graph method to infer the trajectory using single-cell data, and then calculate the geodesic distance as the pseudotime. However, these methods are vulnerable to errors caused by the inferred trajectory. Therefore, the calculated pseudotime suffers from such errors.
Researchers at the University of Nevada, Reno have developed a novel framework for trajectory inference called the single-cell data Trajectory inference method using Ensemble Pseudotime inference (scTEP). scTEP utilizes multiple clustering results to infer robust pseudotime and then uses the pseudotime to fine-tune the learned trajectory. The researchers evaluated the scTEP using 41 real scRNA-seq data sets, all of which had the ground truth development trajectory. They compared the scTEP with state-of-the-art methods using the aforementioned data sets. Experiments on real linear and non-linear data sets demonstrate that their scTEP performed superior on more data sets than any other method. The scTEP also achieved a higher average and lower variance on most metrics than other state-of-the-art methods. In terms of trajectory inference capacity, the scTEP outperforms those methods. In addition, the scTEP is more robust to the unavoidable errors resulting from clustering and dimension reduction.
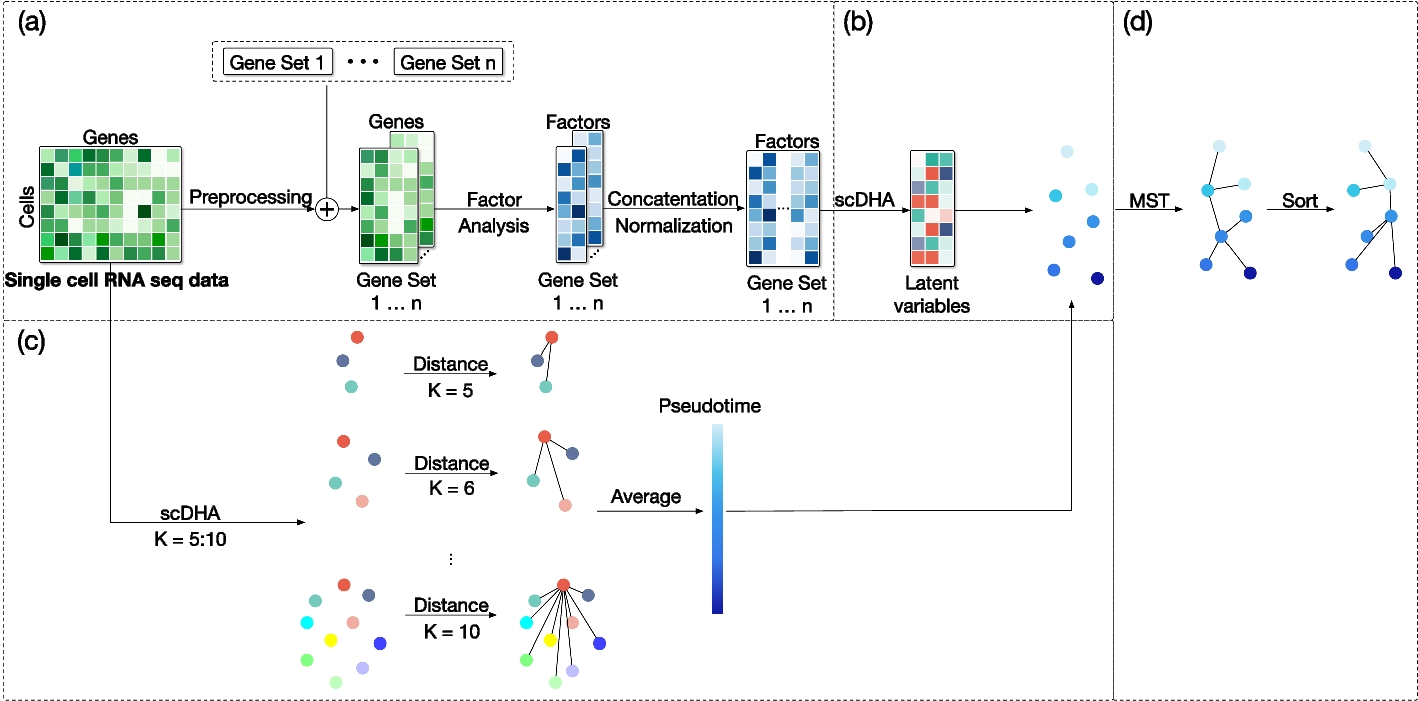
The architecture of the proposed single-cell data Trajectory inference method using Ensemble Pseudotime inference (scTEP)
It consists of four parts: a Data pre-processing and Pathway gene sets intersection, b scDHA clustering and dimension reduction, c Ensemble pseudotime inference, and d Trajectory construction using MST algorithm on clusters and fine-tuned by Pseudotime
The scTEP demonstrates that utilizing multiple clustering results for the pseudotime inference procedure enhances its robustness. Furthermore, robust pseudotime strengthens the accuracy of trajectory inference, which is the most crucial component in the pipeline.
Availability – scTEP is available at https://cran.r-project.org/package=scTEP.
Zhang Y, Tran D, Nguyen T. et al. (2023) A robust and accurate single-cell data trajectory inference method using ensemble pseudotime. BMC Bioinformatics 24, 55. [article]