Complex biological systems are described as a multitude of cell-cell interactions (CCIs). Recent single-cell RNA-sequencing studies focus on CCIs based on ligand-receptor (L-R) gene co-expression but the analytical methods are not appropriate to detect many-to-many CCIs. Researchers at the RIKEN Center for Biosystems Dynamics Research have developed scTensor, a novel method for extracting representative triadic relationships (or hypergraphs), which include ligand-expression, receptor-expression, and related L-R pairs. Through extensive studies with simulated and empirical datasets, the researchers have shown that scTensor can detect some hypergraphs that cannot be detected using conventional CCI detection methods, especially when they include many-to-many relationships.
Implementation of the scTensor package
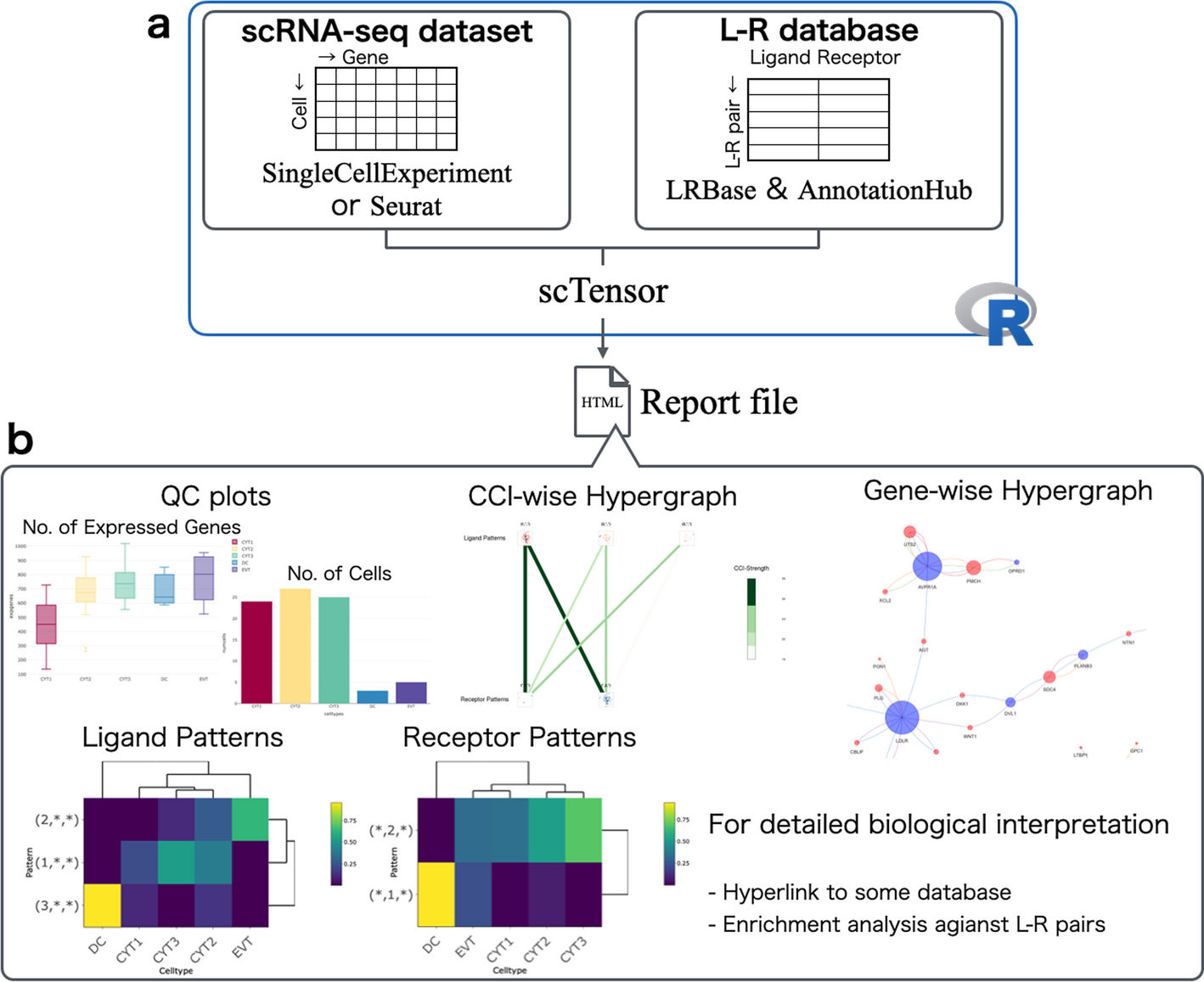
a scTensor is an R package that requires the input of both an scRNA-Seq expression matrix (SingleCellExperiment or Seurat) and a ligand–receptor (L–R) database (LRBase). The LRBase is retrieved from the AnnotationHub remote server, after which a LRBase object is created. b Using these objects, scTensor generates an HTML report file, and the results of cell–cell interaction (CCI) analysis can be visualized with a wide variety of plots
Availability – scTensor is implemented as a freely available R/Bioconductor package: https://bioconductor.org/packages/devel/bioc/html/scTensor.html
Tsuyuzaki K, Ishii M, Nikaido I. (2023) Sctensor detects many-to-many cell-cell interactions from single cell RNA-sequencing data. BMC Bioinformatics 24(1):420. [article]