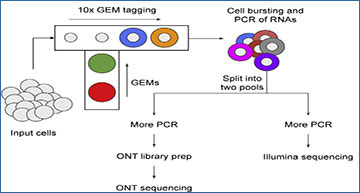
Single-cell RNA sequencing allows for characterizing the gene expression landscape at the cell type level. However, because of its use of short-reads, it is severely limited at detecting full-length features of transcripts such as alternative splicing. New library preparation techniques attempt to extend single-cell sequencing by utilizing both long-reads and short-reads. These techniques split the library material, after it is tagged with cellular barcodes, into two pools: one for short-read sequencing and one for long-read sequencing. However, the challenge of utilizing these techniques is that they require matching the cellular barcodes sequenced by the erroneous long-reads to the cellular barcodes detected by the short-reads.
To overcome this challenge, researchers at the University of British Columbia have developed scTagger, a computational method to match cellular barcodes data from long-reads and short-reads. The researchers tested scTagger against another state-of-the-art tool on both real and simulated datasets, and we demonstrate that scTagger has both significantly better accuracy and time efficiency.
Ebrahimi G, Orabi B, Robinson M, Chauve C, Flannigan R, Hach F. (2022) Fast and accurate matching of cellular barcodes across short-reads and long-reads of single-cell RNA-seq experiments. iScience 25(7):104530. [article]