Single-cell RNA sequencing (scRNA-seq) strives to capture cellular diversity with higher resolution than bulk RNA sequencing. Clustering analysis is critical to transcriptome research as it allows for further identification and discovery of new cell types. Unsupervised clustering cannot integrate prior knowledge where relevant information is widely available. Purely unsupervised clustering algorithms may not yield biologically interpretable clusters when confronted with the high dimensionality of scRNA-seq data and frequent dropout events, which makes identification of cell types more challenging.
Researchers at Xinjiang University have developed scSemiAAE, a semi-supervised clustering model for scRNA sequence analysis using deep generative neural networks. Specifically, scSemiAAE carefully designs a ZINB adversarial autoencoder-based architecture that inherently integrates adversarial training and semi-supervised modules in the latent space. In a series of experiments on scRNA-seq datasets spanning thousands to tens of thousands of cells, scSemiAAE can significantly improve clustering performance compared to dozens of unsupervised and semi-supervised algorithms, promoting clustering and interpretability of downstream analyses.
The illustration of scSemiAAE model
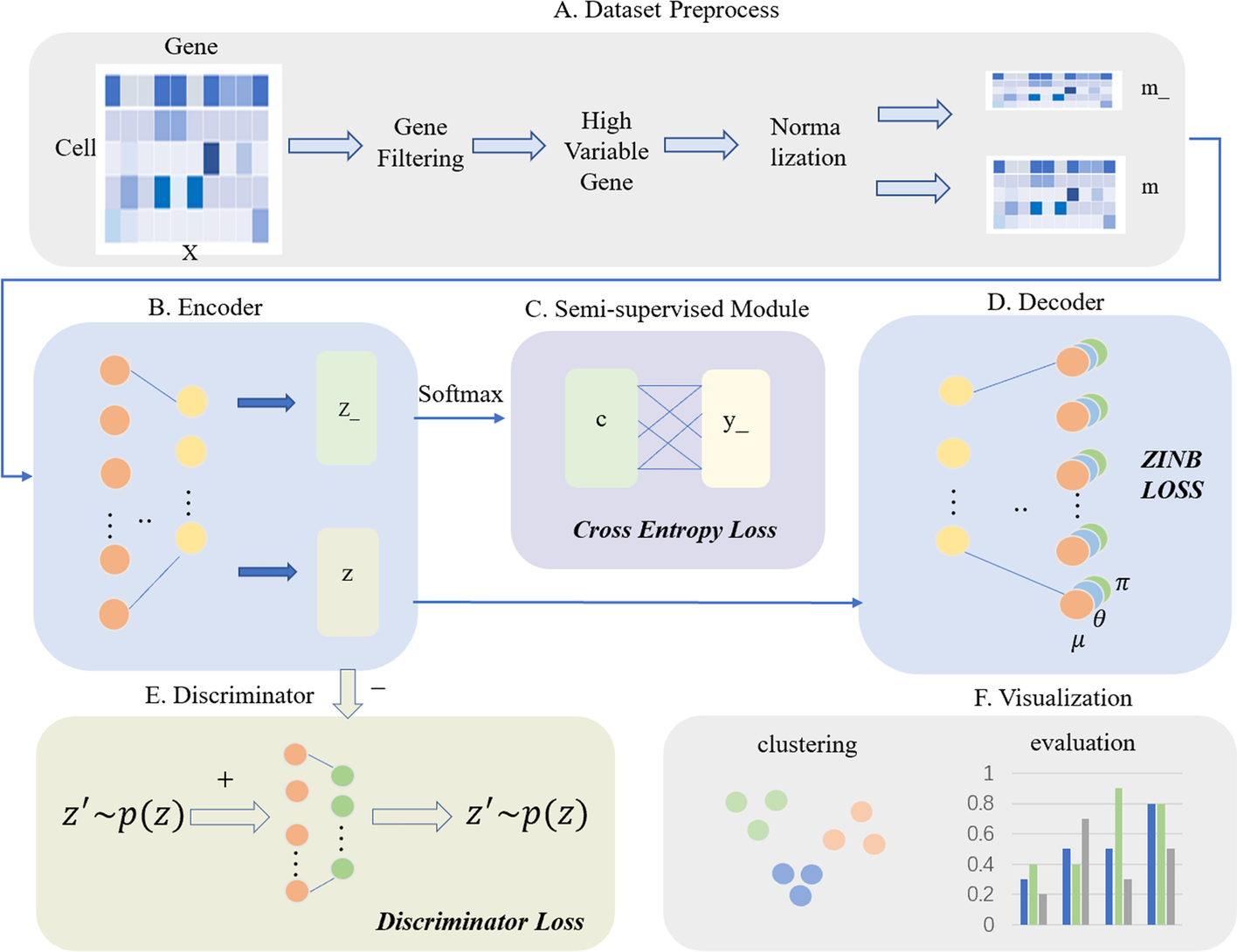
A The scRNA-seq count matrix X is preprocessed through gene filtering, screening of highly variable genes, and normalization. Next, it is divided into m_ and m depending on whether it contains true labels. B The encoder receives m_ and m to generate the corresponding latent variables z_ and z, respectively. C The SoftMax layer transforms the latent vector z_ into the pseudo-label c, which is then combined with the partial true label y_ to create a cross-entropy loss. D The decoder reconstructs the potential representation z with a zero-inflated negative binomial loss constraint. E Simultaneously, the latent feature z is fed to the discriminator for adversarial training, comprising the discriminator loss. F After completing training process, all the latent z and labels c are concatenated, and the final clustering results are given by a Gaussian mixture model
Availability – The tool is available from https://github.com/WHang98/scSemiAAE .
Wang Z, Wang H, Zhao J, Zheng C. (2023) scSemiAAE: a semi-supervised clustering model for single-cell RNA-seq data. BMC Bioinformatics 24(1):217. [article]