Cell clustering is a prerequisite for identifying differentially expressed genes (DEGs) in single-cell RNA sequencing (scRNA-seq) data. Obtaining a perfect clustering result is of central importance for subsequent analyses, but not easy. Additionally, the increase in cell throughput due to the advancement of scRNA-seq protocols exacerbates many computational issues, especially regarding method runtime. To address these difficulties, a new, accurate, and fast method for detecting DEGs in scRNA-seq data is needed.
Researchers at Xidian University have developed single-cell minimum enclosing ball (scMEB), a novel and fast method for detecting single-cell DEGs without prior cell clustering results. The proposed method utilizes a small part of known non-DEGs (stably expressed genes) to build a minimum enclosing ball and defines the DEGs based on the distance of a mapped gene to the center of the hypersphere in a feature space.
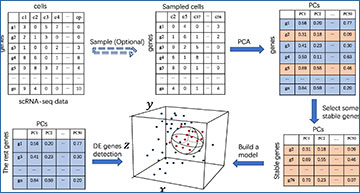
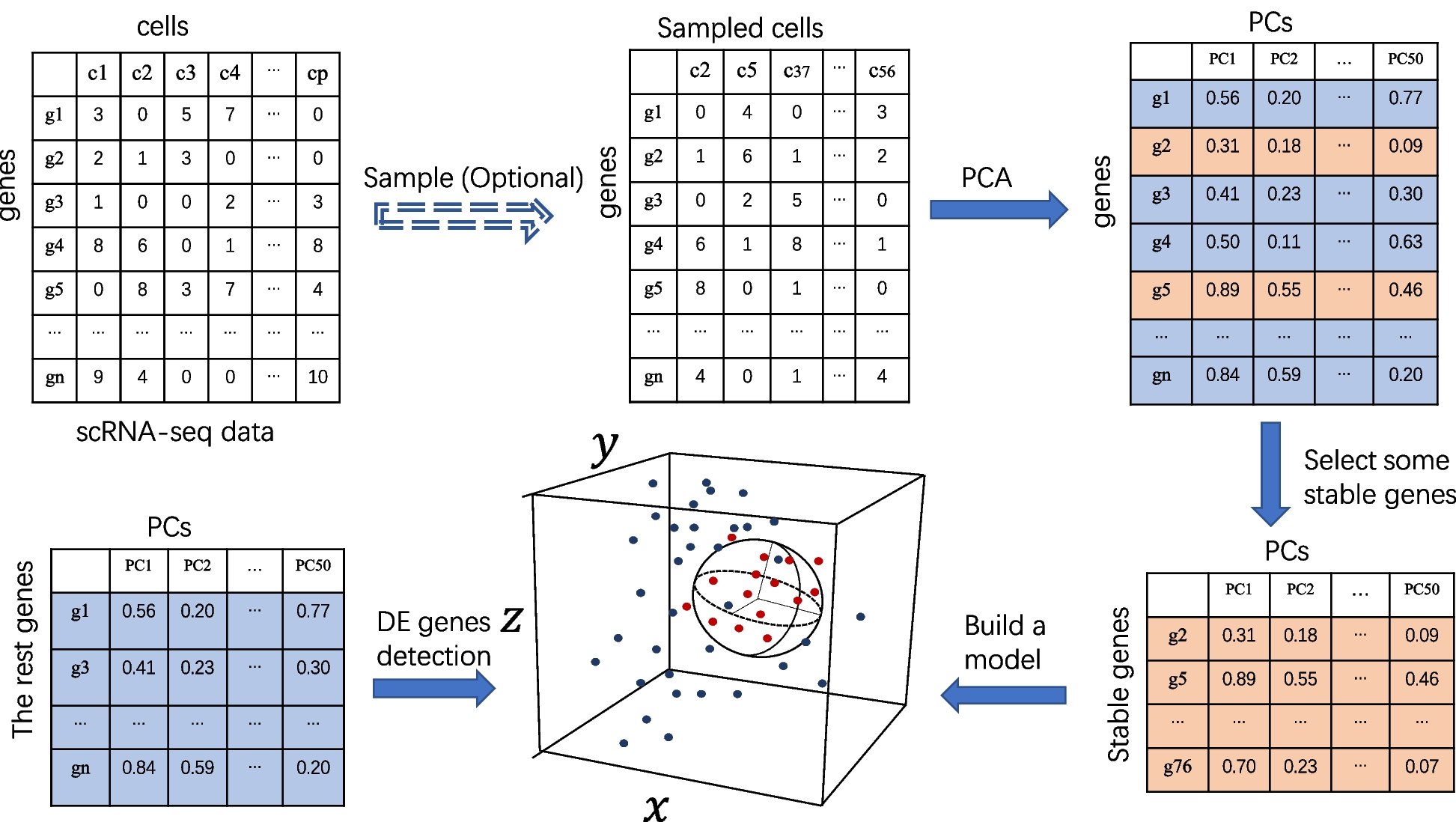
Overview of the scMEB workflow
Randomly sampling some important cells before PCA is optional, and the purpose is to speed up the calculation. The first 50 PCs could be obtained from the sampled/unsampled cells. A small part of the stable genes was used to build the model. A sphere that encloses the training data in the feature space was found. The remaining genes were divided into DEGs or non-DEGs according to whether they were outside or inside the enclosing ball, respectively
The researchers compared scMEB to two different approaches that could be used to identify DEGs without cell clustering. The investigation of 11 real datasets revealed that scMEB outperformed rival methods in terms of cell clustering, predicting genes with biological functions, and identifying marker genes. Moreover, scMEB was much faster than the other methods, making it particularly effective for finding DEGs in high-throughput scRNA-seq data.
Availability – a package scMEB for the proposed available at: https://github.com/FocusPaka/scMEB.
Zhu J, Yang Y. (2023) scMEB: a fast and clustering-independent method for detecting differentially expressed genes in single-cell RNA-seq data. BMC Genomics 24(1):280. [article]