Class-switch recombination (CSR) is an integral part of B cell maturation. Researchers at the University College London have developed sciCSR (pronounced ‘scissor’, single-cell inference of class-switch recombination), a computational pipeline that analyzes CSR events and dynamics of B cells from single-cell RNA sequencing (scRNA-seq) experiments. Validated on both simulated and real data, sciCSR re-analyzes scRNA-seq alignments to differentiate productive heavy-chain immunoglobulin transcripts from germline ‘sterile’ transcripts. From a snapshot of B cell scRNA-seq data, a Markov state model is built to infer the dynamics and direction of CSR.
The sciCSR pipeline to infer B cell state transitions
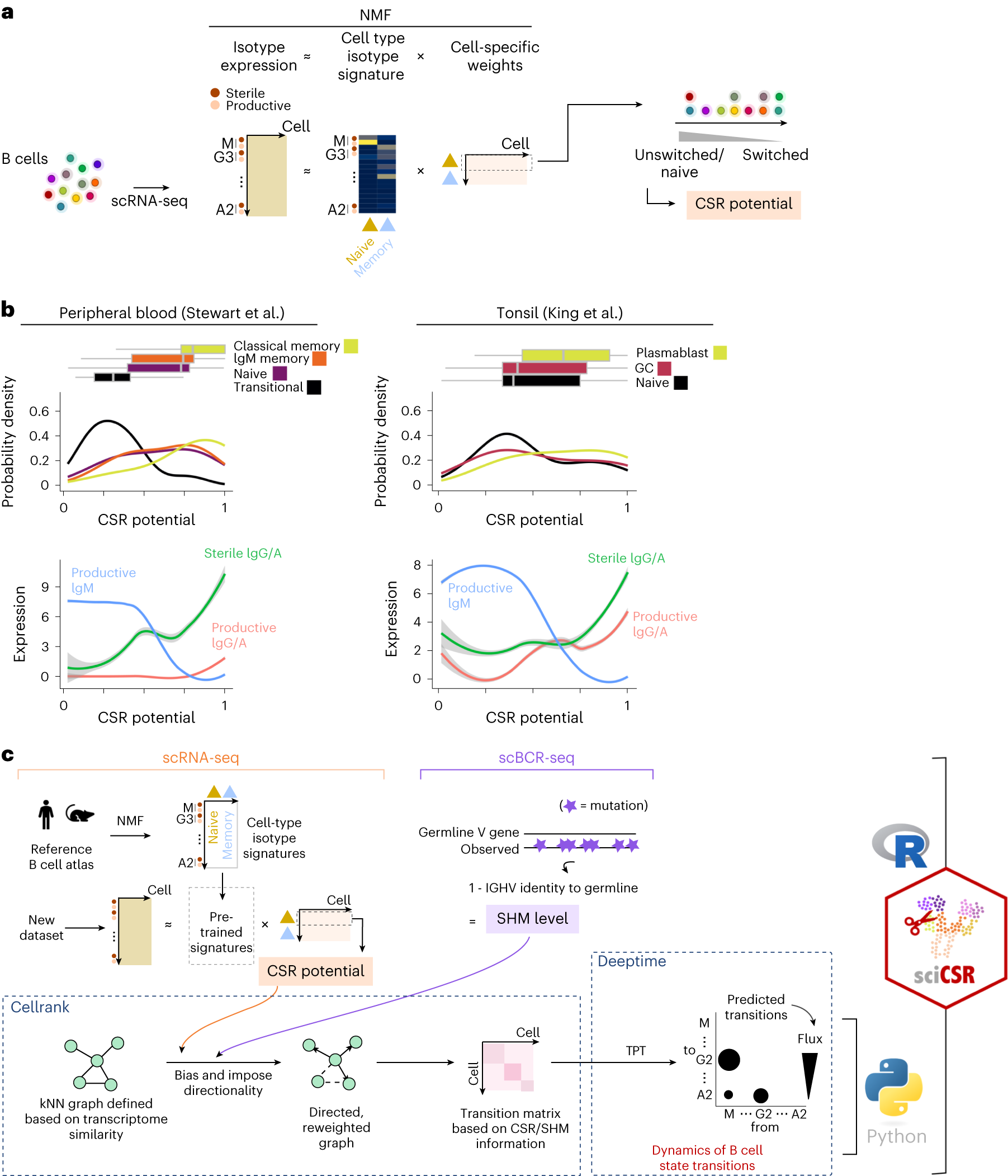
a, Reference B cell atlases from mouse and human were subjected to NMF to extract isotype signatures that describe naive/memory B cell subsets. The naive signature was used to rank the cells by their maturation status, to generate a score termed ‘CSR potential’. b, Top: distribution of CSR potential score for B cell subsets in the Stewart et al. peripheral blood (left) and King et al. tonsil (right) datasets. Boxplots depict distribution of the CSR potential scores, with vertical lines indicating lower quartile, median and upper quartile of the distributions. Bottom: expression level of different productive and sterile IgH transcripts along the axis of the inferred CSR potentials in the datasets. c, Schematic to illustrate the sciCSR workflow for inferring state transitions.
Applying sciCSR on severe acute respiratory syndrome coronavirus 2 vaccination time-course scRNA-seq data, the researchers observe that sciCSR predicts, using data from an earlier time point in the collected time-course, the isotype distribution of B cell receptor repertoires of subsequent time points with high accuracy (cosine similarity ~0.9). Using processes specific to B cells, sciCSR identifies transitions that are often missed by conventional RNA velocity analyses and can reveal insights into the dynamics of B cell CSR during immune response.
Availability – Source code for the sciCSR package is available at https://github.com/Fraternalilab/sciCSR.
Ng JCF, Montamat Garcia G, Stewart AT, Blair P, Mauri C, Dunn-Walters DK, Fraternali F. (2023) sciCSR infers B cell state transition and predicts class-switch recombination dynamics using single-cell transcriptomic data. Nat Methods [Epub ahead of print]. [article]