Extrachromosomal DNAs (ecDNAs) are common in cancer, but many questions about their origin, structural dynamics and impact on intratumor heterogeneity are still unresolved. Researchers from the Universitätsmedizin Berlin have developed single-cell extrachromosomal circular DNA and transcriptome sequencing (scEC&T-seq), a method for parallel sequencing of circular DNAs and full-length mRNA from single cells. By applying scEC&T-seq to cancer cells, the researchers describe intercellular differences in ecDNA content while investigating their structural heterogeneity and transcriptional impact. Oncogene-containing ecDNAs were clonally present in cancer cells and drove intercellular oncogene expression differences. In contrast, other small circular DNAs were exclusive to individual cells, indicating differences in their selection and propagation. Intercellular differences in ecDNA structure pointed to circular recombination as a mechanism of ecDNA evolution. These results demonstrate scEC&T-seq as an approach to systematically characterize both small and large circular DNA in cancer cells, which will facilitate the analysis of these DNA elements in cancer and beyond.
scEC&T-seq enables enrichment and detection of circular DNA in single cells
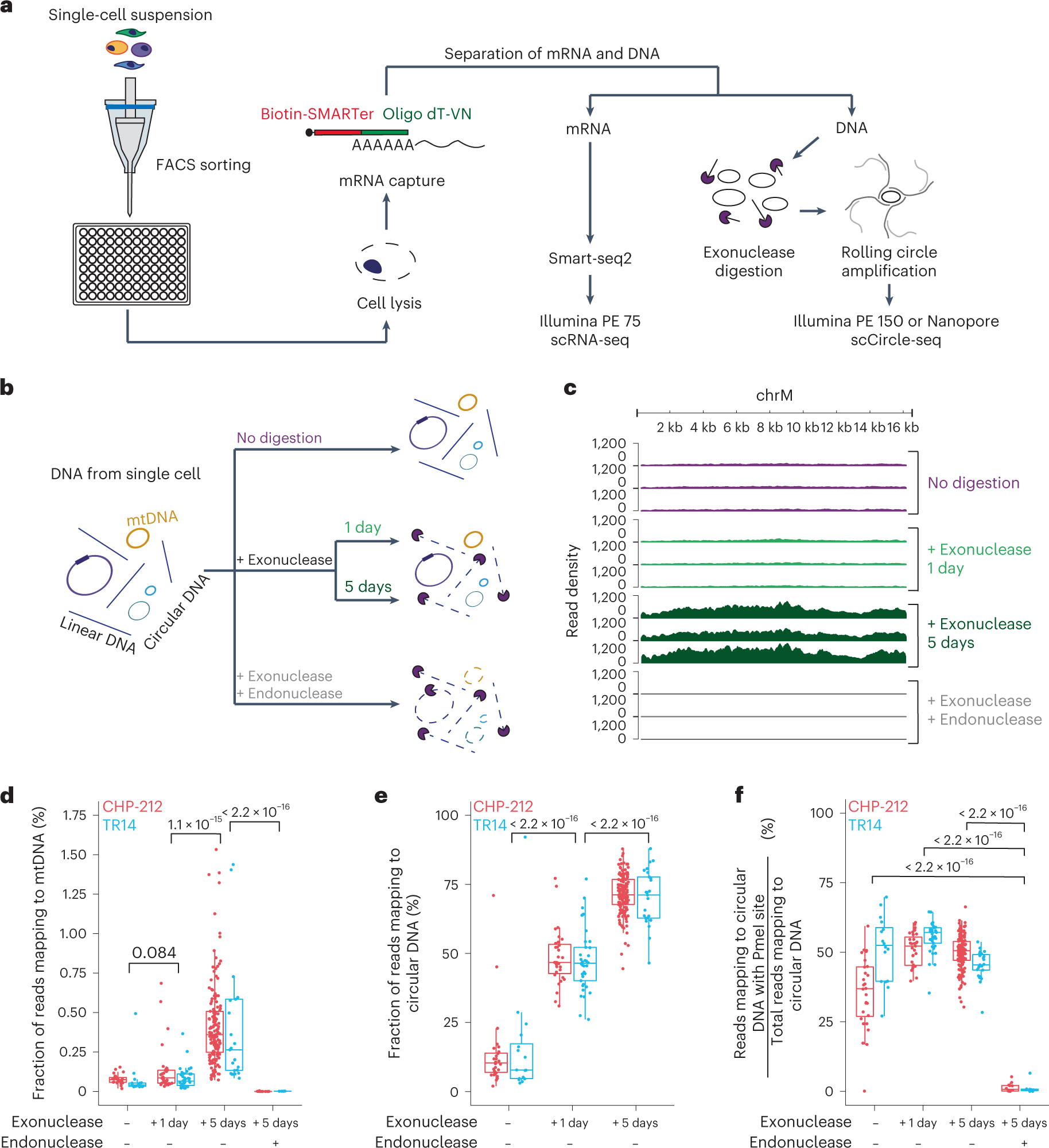
a, Schematic of the scEC&T-seq method. b, Schematic representation of the experimental conditions and expected outcomes. c, Genome tracks comparing read densities on mtDNA (chrM) in three exemplary CHP-212 cells for each experimental condition tested. Top to bottom, No digestion (purple), 1-day exonuclease digestion (light green), 5-day exonuclease digestion (dark green) and endonuclease digestion with PmeI before 5-day exonuclease digestion (gray). d, Fraction of sequencing reads mapping to mtDNA in each experimental condition in CHP-212 (red) and TR14 (blue) cells. e, Fraction of sequencing reads mapping to circular DNA regions identified by scEC&T-seq in each experimental condition in CHP-212 and TR14 cells. f, Fraction of sequencing reads mapping to circular DNA regions with the endonuclease PmeI targeting the sequence identified by scEC&T-seq in each experimental condition in CHP-212 and TR14 cells. d–f, Sample size is identical across conditions: no digestion (n = 16 TR14 cells, n = 28 CHP-212 cells); 1-day exonuclease digestion (n = 37 TR14 cells, n = 31 CHP-212 cells); 5-day exonuclease digestion (n = 25 TR14 cells, n = 150 CHP-212 cells); and endonuclease digestion with PmeI before 5-day exonuclease digestion (n = 6 TR14 cells, n = 12 CHP-212 cells). All statistical analyses correspond to a two-sided Welch’s t-test. P values are shown. In all boxplots, the boxes represent the 25th and 75th percentiles with the center bar as the median value and the whiskers representing the furthest outlier ≤1.5× the interquartile range (IQR) from the box.
Chamorro González R, Conrad T, Stöber MC, Xu R, Giurgiu M, Rodriguez-Fos E, Kasack K, Brückner L, van Leen E, Helmsauer K, Dorado Garcia H, Stefanova ME, Hung KL, Bei Y, Schmelz K, Lodrini M, Mundlos S, Chang HY, Deubzer HE, Sauer S, Eggert A, Schulte JH, Schwarz RF, Haase K, Koche RP, Henssen AG. (2023) Parallel sequencing of extrachromosomal circular DNAs and transcriptomes in single cancer cells. Nat Genet [Epub ahead of print]. [article]