Single-cell RNA-sequencing (scRNA-seq) has been widely used for disease studies, where sample batches are collected from donors under different conditions including demographic groups, disease stages, and drug treatments. It is worth noting that the differences among sample batches in such a study are a mixture of technical confounders caused by batch effect and biological variations caused by condition effect. However, current batch effect removal methods often eliminate both technical batch effect and meaningful condition effect, while perturbation prediction methods solely focus on condition effect, resulting in inaccurate gene expression predictions due to unaccounted batch effect.
Researchers at the Georgia Institute of Technology have developed scDisInFact, a deep learning framework that models both batch effect and condition effect in scRNA-seq data. scDisInFact learns latent factors that disentangle condition effect from batch effect, enabling it to simultaneously perform three tasks: batch effect removal, condition-associated key gene detection, and perturbation prediction. The researchers evaluated scDisInFact on both simulated and real datasets, and compared its performance with baseline methods for each task. Their results demonstrate that scDisInFact outperforms existing methods that focus on individual tasks, providing a more comprehensive and accurate approach for integrating and predicting multi-batch multi-condition single-cell RNA-sequencing data.
Overview of scDisInFact
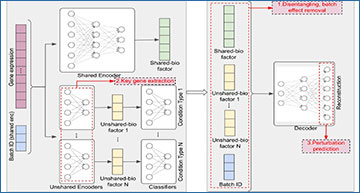
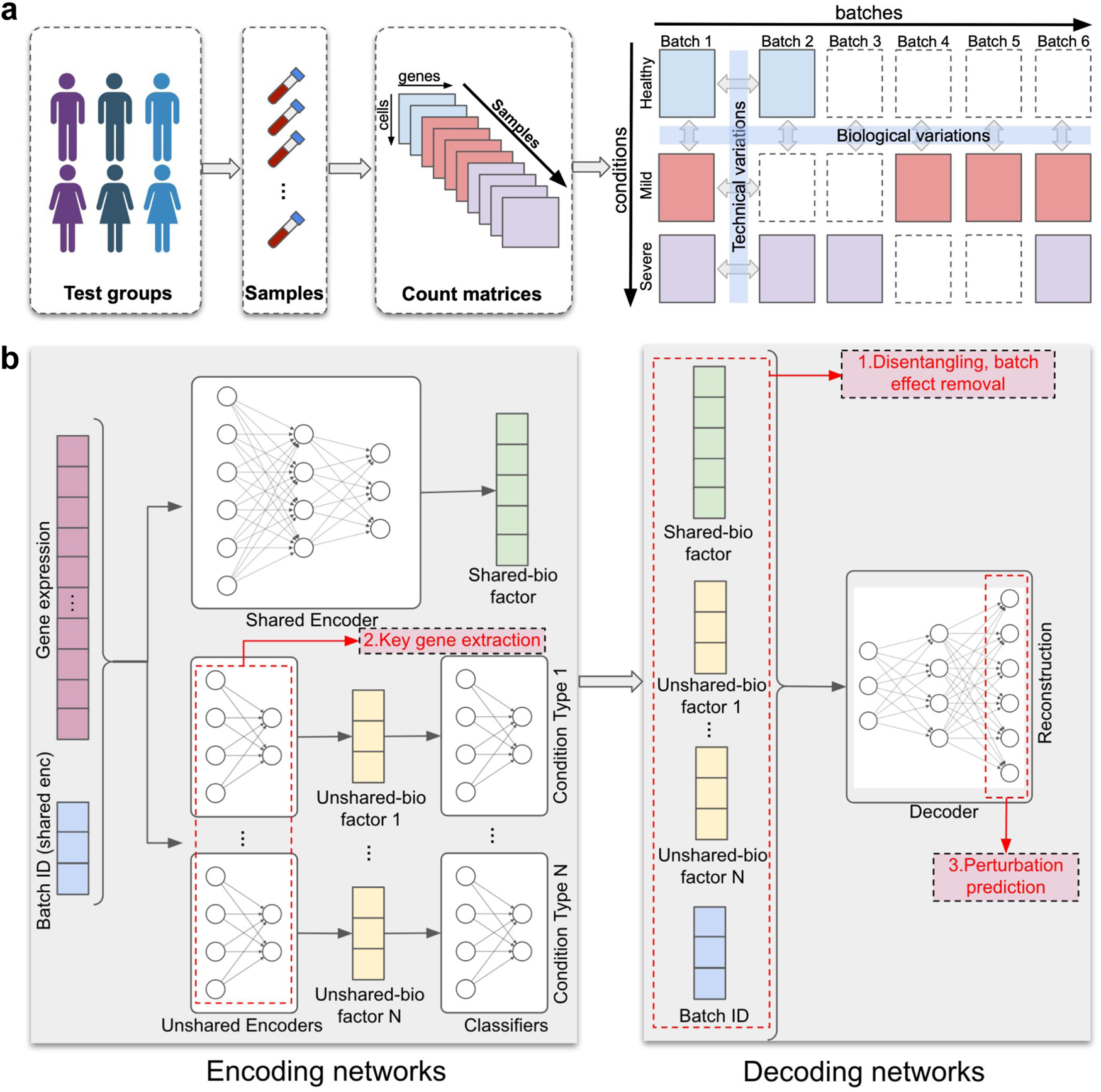
a scDisInFact is applied on multi-batch multi-condition datasets where count matrices from disease studies are obtained from different experimental batches and conditions. Human figure created with BioRender.com. b The neural network structure of scDisInFact. scDisInFact uses an encoding network (left) to learn the disentangled latent factors, and uses a decoding network (right) to generate gene expression data from the latent factors. It is designed for tasks including (1) batch effect removal (latent factors disentanglement), (2) condition-associated key genes detection, and (3) perturbation prediction. Neural network illustration adapted from LeNail.
Availability – The code of scDisInFact is available on GitHub with the link: https://github.com/ZhangLabGT/scDisInFact.
Zhang Z, Zhao X, Bindra M, Qiu P, Zhang X. (2024) scDisInFact: disentangled learning for integration and prediction of multi-batch multi-condition single-cell RNA-sequencing data. Nat Commun 15(1):912. [article]