RNA molecules are emerging as key players in cellular processes, going beyond their traditional role as mere messengers of genetic information. These molecules are now recognized for their active involvement in various cellular pathways, driven by their complex secondary structures.
Understanding RNA secondary structures is crucial as it provides insights into how these molecules function within cells. However, traditional experimental methods for determining RNA structures are costly and time-consuming, making large-scale investigations challenging. To overcome this hurdle, researchers have turned to computational tools to predict RNA structures from their sequences.
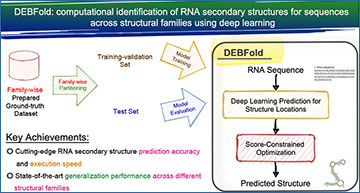
Recent advancements in deep learning have opened new avenues for RNA structure prediction. However, existing deep-learning-based tools face challenges such as model overfitting and limited applicability across different structural families. In response, a team of researchers at National Cheng Kung University, Taiwan has developed a groundbreaking approach called DEBFold (deep ensemble boosting and folding).
Overview of DEBFold
Notations: Conv─convolution layers; FC─fully connected layers; TransConv─transpose convolution layers; MFE: minimum free energy.
DEBFold utilizes a two-stage prediction strategy that incorporates convolution encoding/decoding and self-attention mechanisms, enhancing existing thermodynamic structure models. This innovative approach not only improves prediction accuracy but also ensures robust generalization across different RNA structural families.
The key to DEBFold’s success lies in its rigorous model training process, which addresses issues such as overfitting and ensures reliable predictions. In comparative tests using family-wise reserved datasets and PDB-derived test sets, DEBFold outperformed traditional tools and existing deep-learning methods, demonstrating its superior performance in RNA structure prediction.
DEBFold represents a significant breakthrough in RNA structure prediction technology. Its cutting-edge deep-learning-based approach offers unparalleled accuracy and generalization across diverse RNA sequences. With DEBFold, researchers have a powerful tool at their disposal for unraveling the hidden structures of RNA molecules, paving the way for groundbreaking discoveries in molecular biology and potential applications in drug development and disease treatment.
Availability – The DEBFold tool can be accessed at https://cobis.bme.ncku.edu.tw/DEBFold/.
Yang TH. (2024) DEBFold: Computational Identification of RNA Secondary Structures for Sequences across Structural Families Using Deep Learning. J Chem Inf Model [Epub ahead of print]. [article]