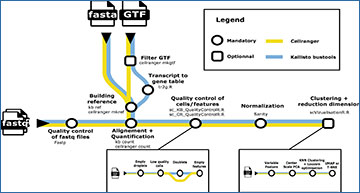
Single cell transcriptomics has recently seen a surge in popularity, leading to the need for data analysis pipelines that are reproducible, modular, and interoperable across different systems and institutions. To meet this demand, researchers at the CNRS introduce scAN1.0, a processing pipeline for analyzing 10X single cell RNA sequencing data. scAN1.0 is built using the Nextflow DSL2 and can be run on most computational systems. The modular design of Nextflow pipelines enables easy integration and evaluation of different blocks for specific analysis steps. The researchers demonstrate the usefulness of scAN1.0 by showing its ability to examine the impact of the mapping step during the analysis of two datasets: (i) a 10X scRNAseq of a human pituitary gonadotroph tumor dataset and (ii) a murine 10X scRNAseq acquired on CD8 T cells during an immune response.
A metromap view of the scAN1.0 pipeline. Tools used for the processing steps are given below the name of the process.
Lepetit M, Ilie MD, Chanal M, Raverot G, Bertolino P, Arpin C, Picard F, Gandrillon O. (2023) scAN1.0: A reproducible and standardized pipeline for processing 10X single cell RNAseq data. In Silico Biol 15(1-2):11-21. [article]