Single-cell RNA sequencing (scRNA-seq) has emerged as a powerful technique to decipher tissue composition at the single-cell level and to inform on disease mechanisms, tumor heterogeneity, and the state of the immune microenvironment. Although multiple methods for the computational analysis of scRNA-seq data exist, their application in a clinical setting demands standardized and reproducible workflows, targeted to extract, condense, and display the clinically relevant information. To this end, researchers at ETH Zurich designed scAmpi (Single Cell Analysis mRNA pipeline), a workflow that facilitates scRNA-seq analysis from raw read processing to informing on sample composition, clinically relevant gene and pathway alterations, and in silico identification of personalized candidate drug treatments. The researchers demonstrate the value of this workflow for clinical decision making in a molecular tumor board as part of a clinical study.
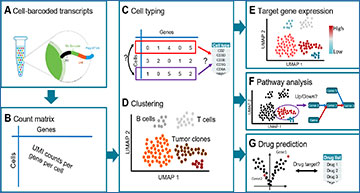
Overview of the workflow implemented in scAmpi,
showing a tumor sample analysis as an example
Starting from droplet-based 10x Genomics raw data (A), genome-wide read counts for each cell are generated (B). This gene-by-cell count matrix is the basis for cell type prediction (C) and unsupervised clustering (D) to determine the cell type composition and tumor heterogeneity. Subsequent steps include gene expression (E) and gene set (F) analysis, and drug candidate identification (G).
Bertolini A, Prummer M, Tuncel MA, Menzel U, Rosano-González ML, Kuipers J, et al. (2022) scAmpi—A versatile pipeline for single-cell RNA-seq analysis from basics to clinics. PLoS Comput Biol 18(6): e1010097. [article]