When biologically interpretation of the data obtained from the single-cell RNA sequencing (scRNA-seq) analysis is attempted, additional information on the location of the single cells, behavior of the surrounding cells, and the microenvironment they generate, would be very important. University of Tokyo researchers have developed an inexpensive, high throughput application while preserving spatial organization, named “semibulk RNA-seq” (sbRNA-seq). The researchers utilized a microfluidic device specifically designed for the experiments to encapsulate both a barcoded bead and a cell aggregate (a semibulk) into a single droplet. Using sbRNA-seq, they first analyzed mouse kidney specimens. In the mouse model, they could associate the pathological information with the gene expression information. The researchers validated the results using spatial transcriptome analysis and found them highly consistent. When they applied the sbRNA-seq analysis to the human breast cancer specimens, they identified spatial interactions between a particular population of immune cells and that of cancer-associated fibroblast cells, which were not precisely represented solely by the single-cell analysis. Semibulk analysis may provide a convenient and versatile method, compared to a standard spatial transcriptome sequencing platform, to associate spatial information with transcriptome information.
Muto K, Tsuchiya I, Kim SH. et al. (2022) Semibulk RNA-seq analysis as a convenient method for measuring gene expression statuses in a local cellular environment. Sci Rep [Epub ahead of print]. [article]
A workflow of sbRNA-seq
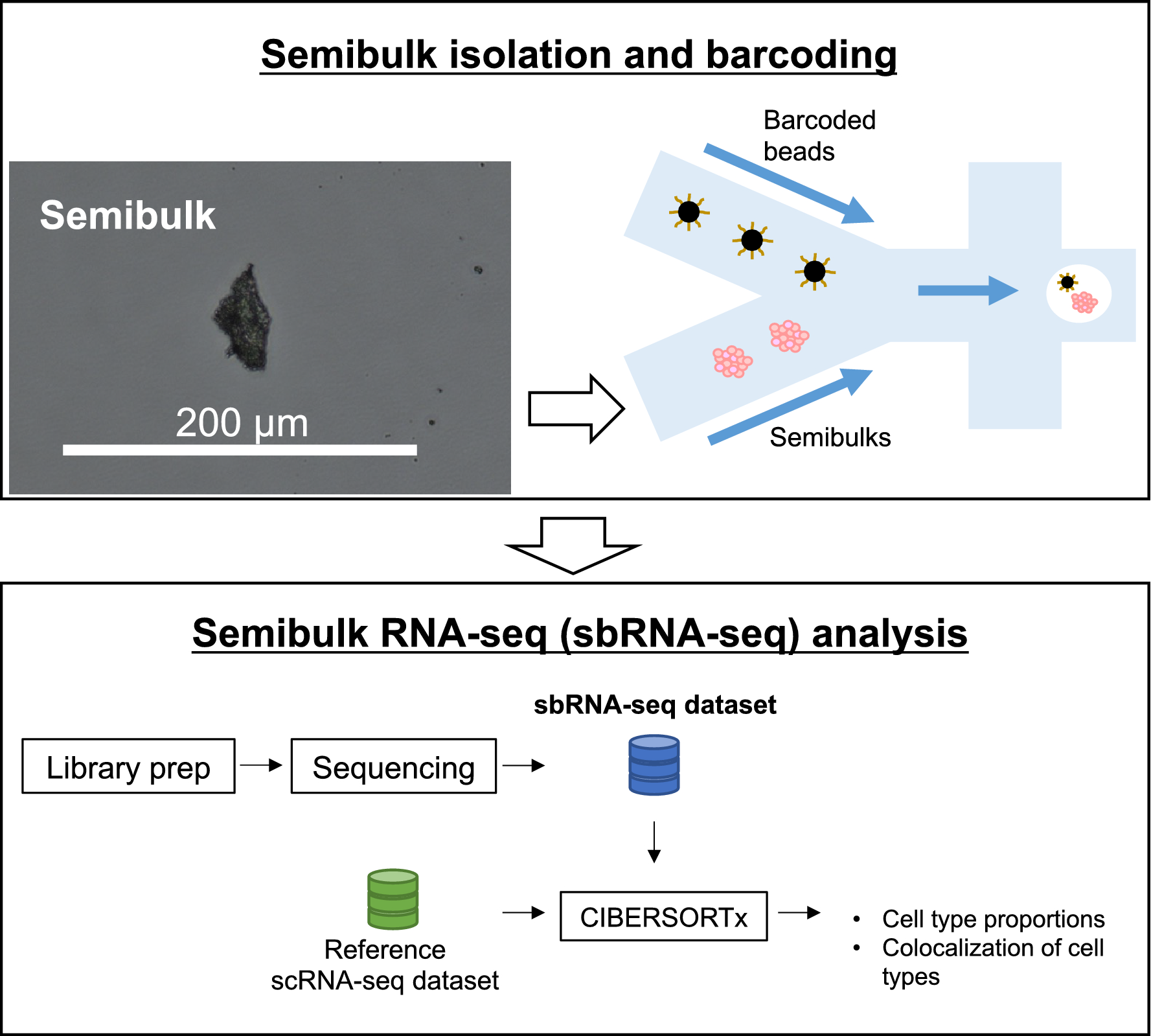 Overall schematic representation. Cell aggregates from tissues are isolated and mixed with a barcoded bead and reaction solution within a droplet. In the droplet, reverse transcription is conducted to generate barcoded cDNA. Library preparation and sequencing are performed to generate sbRNA-seq data. After sequencing, deconvolution analysis is conducted using CIBERSORTx to estimate the cell-type proportion of each semibulk using scRNA-seq data as a reference.
Overall schematic representation. Cell aggregates from tissues are isolated and mixed with a barcoded bead and reaction solution within a droplet. In the droplet, reverse transcription is conducted to generate barcoded cDNA. Library preparation and sequencing are performed to generate sbRNA-seq data. After sequencing, deconvolution analysis is conducted using CIBERSORTx to estimate the cell-type proportion of each semibulk using scRNA-seq data as a reference.
Muto K, Tsuchiya I, Kim SH. et al. (2022) Semibulk RNA-seq analysis as a convenient method for measuring gene expression statuses in a local cellular environment. Sci Rep [Epub ahead of print]. [article]