Single-cell transcriptional profiling reveals cell heterogeneity and clinically relevant traits in intra-operatively collected patient-derived tissue. So far, single-cell studies have been constrained by the requirement for prospectively collected fresh or cryopreserved tissue. This limitation might be overcome by recent technical developments enabling single-cell analysis of FFPE tissue.
Researchers at the Universitätsmedizin Berlin found that fresh tissue and FFPE routine blocks can be employed for the robust detection of clinically relevant traits on the single-cell level. Specifically, single-cell maps of fresh patient tissues and corresponding FFPE tissue blocks could be integrated into common low-dimensional representations, and cell subtype clusters showed highly correlated transcriptional strengths of signaling pathway, hallmark, and clinically useful signatures, although expression of single genes varied due to technological differences. FFPE tissue blocks revealed higher cell diversity compared to fresh tissue. In contrast, single-cell profiling of cryopreserved tissue was prone to artifacts in the clinical setting.
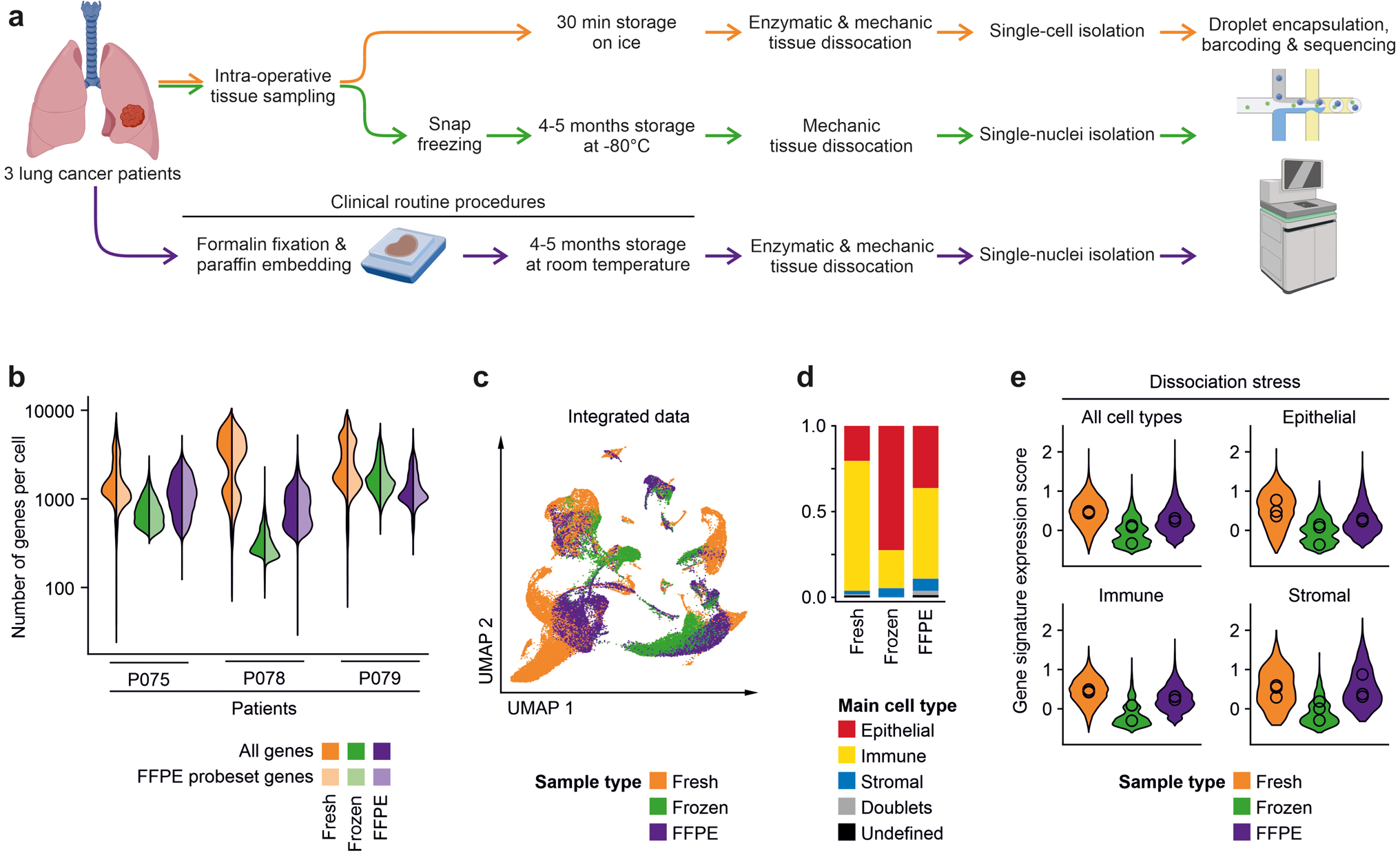
Workflow and quality metrics of fresh, frozen and FFPE tissue single-cell analysis
a Workflow of tissue specimens used for the study. In short, fresh tissues were procured intra-operatively, dissociated enzymatically, and cell suspensions were used for single-cell library preparation. Cryopreserved tissues were stored for 4–5 months at −80 °C, homogenized in the frozen state for nuclei isolation, and nucleus suspensions were used for library preparation. For FFPE analysis, 4–5 months old routine FFPE blocks were dissociated enzymatically and mechanically, and nucleus suspensions were used for library preparation. b Numbers of genes called per cell in the various libraries. Full colors: all genes; lighter colors: genes limited by FFPE probe set. c UMAPs based on the top 10 principal components of all single-cell transcriptomes after filtering and data integration, color-coded by fresh, frozen or FFPE tissue origin. d Quantification of main cell type, by clustering and calling of cell type-specific marker genes. e Module score of gene signatures related to dissociation stress in the various main cell types, by fresh, frozen and FFPE origin, mean scores per patient indicated by circles
This analysis highlights the potential of single-cell profiling in the analysis of retrospectively and prospectively collected archival pathology cohorts and increases the applicability in translational research.
Trinks A, Milek M, Beule D, Kluge J, Florian S, Sers C, Horst D, Morkel M, Bischoff P. (2024) Robust detection of clinically relevant features in single-cell RNA profiles of patient-matched fresh and formalin-fixed paraffin-embedded (FFPE) lung cancer tissue. Cell Oncol (Dordr) [Epub ahead of print]. [article]