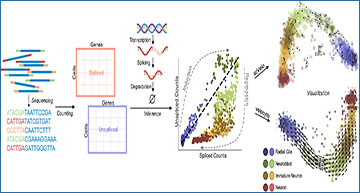
Single-cell sequencing data are snapshots of biological processes, making it challenging to infer dynamic relationships between cell types. RNA velocity attempts to bypass this challenge by treating the unspliced RNA content as a proxy for spliced RNA content in the near future, and using this “extrapolation” to build directional relationships. However, the method, as implemented in several software packages, is not yet reliable enough to be actionable, in part due to the large number of arbitrary, user-set hyperparameters, as well as fundamental incompatibilities between the biophysics of transcription in the living cell and the models used throughout the velocity workflows.
Researchers at the California Institute of Technology discuss these issues, and use existing results from the fields of stochastic modeling and fluorescence transcriptomics to develop an alternative theoretical framework. They show that their framework can facilitate the development and inference of physically consistent models for sequencing data, as well as the unification of single-cell analyses to self-consistently treat variation due to cell type dynamics and identities, the stochasticity inherent to single-molecule processes, and the uncertainty introduced by sequencing experiments.
An RNA velocity workflow, beginning with read processing and ending with two-dimensional projection, and the parameters that must be specified by the user.
Gorin G, Fang M, Chari T, Pachter L (2022) RNA velocity unraveled. PLoS Comput Biol 18(9): e1010492. [article]