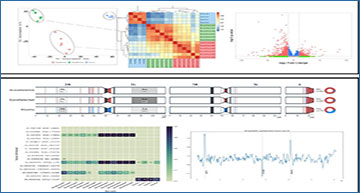
Researchers at the Liverpool School of Tropical Medicine have developed a reproducible and scalable Snakemake workflow, called RNA-Seq-Pop, which provides end-to- end analysis of RNA-Seq data sets. The workflow allows the user to perform quality control, differential expression analyses, call genomic variants and generate a range of summary statistics. Additional options include the calculation of allele frequencies of variants of interest, summaries of genetic variation and population structure (in measures such as nucleotide diversity, Watterson’s θ, and PCA), and genome wide selection scans (Fst, PBS), together with clear visualisations. The researchers demonstrate the utility of the workflow by investigating pyrethroid-resistance in selected strains of the major malaria mosquito, Anopheles gambiae. The workflow provides additional modules specifically for An. gambiae, including estimating recent ancestry and determining the karyotype of common chromosomal inversions.
The RNA-Seq-Pop workflow and example outputs
The workflow has been designed for ease of use, requiring only a configuration file to set up workflow choices and a sample sheet to provide sample metadata. Modules highlighted in green are specific to An. gambiae s.l.
RNA-Seq-Pop is designed for ease of use, does not require programming skills and integrates the package manager Conda to ensure that all dependencies are automatically installed for the user. The researchers anticipate that the workflow will provide a useful tool to facilitate reproducible, transcriptomic studies in An. gambiae and other taxa.
Nagi SC, Oruni A, Weetman D, Donnelly MJ. (2022) RNA-Seq-Pop: Exploiting the sequence in RNA-Seq – a Snakemake workflow reveals patterns of insecticide resistance in the malaria vector Anopheles gambiae. bioRXiv [online preprint]. [article]