The heterogeneous composition of cellular transcriptomes poses a major challenge for detecting weakly expressed RNA classes, as they can be obscured by abundant RNAs. Although biochemical protocols can enrich or deplete specified RNAs, they are time-consuming, expensive and can compromise RNA integrity. Researchers at the Australian National University have developed RISER, a biochemical-free technology for the real-time enrichment or depletion of RNA classes. RISER performs selective rejection of molecules during direct RNA sequencing by identifying RNA classes directly from nanopore signals with deep learning and communicating with the sequencing hardware in real time. RISER achieved a 4x read (6x nucleotide) enrichment of long non-coding RNAs during live sequencing by depleting the dominant messenger (mRNA) and mitochondrial RNA classes, and a 4x read (5x nucleotide) enrichment of non-globin mRNA in whole blood by depleting globin mRNA. Using a GPU or a CPU, RISER is faster than GPU-accelerated basecalling and mapping. RISER’s modular and retrainable software and intuitive command-line interface allow easy adaptation to other RNA classes.
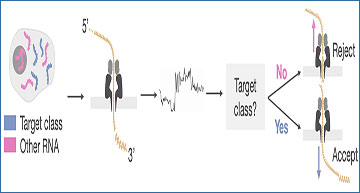
RISER identifies RNA classes from the 3’ end of DRS signals
a, RISER classifies RNA molecules as they commence sequencing by directly assessing raw nanopore signals, then sends an accept or reject decision to the sequencing hardware depending on the user-defined target RNA class and whether the user wants to enrich or deplete the target class (shown: target depletion). The accepted reads are sequenced to completion, while the rejected reads are truncated. b, Percentage of reads in the training dataset (y-axis) with raw signals long enough to be input to RISER, for each candidate input signal length expressed in seconds (x-axis). c-e, Model performance on the test set for each candidate input signal length (x-axes), color-coded by the three convolutional network architectures assessed: “vanilla” convolutional neural network (CNN) (cyan), residual network (ResNet) (dark blue), temporal convolutional network (TCN) (pink). We show the accuracy (c), ratio of true positive rate (TPR) to false positive rate (FPR) (d) and mean prediction time per batch of signals, expressed in milliseconds (e). f, Neural network architecture for the CNN model selected to implement RISER.
Availability – RISER is available at https://github.com/comprna/riser.
Sneddon A, Ravindran A, Shanmuganandam S, Mdhu Kanchi M, Hein N, Smon Jiang S, Shirokikh N , Eyras E. (2023) Biochemical-free enrichment or depletion of RNA classes in real-time during direct RNA sequencing with RISER. bioRXiv [online preprint]. [article]