With the advancement of technology, we can generate and access large-scale, high dimensional and diverse genomics data, especially through single-cell RNA sequencing (scRNA-seq). However, integrative downstream analysis from multiple scRNA-seq datasets remains challenging due to batch effects.
Researchers from Yale University have now developed a light-structured deep learning framework called ResPAN for scRNA-seq data integration. ResPAN is based on Wasserstein Generative Adversarial Network (WGAN) combined with random walk mutual nearest neighbor pairing and fully skip-connected autoencoders to reduce the differences among batches. The researchers also discuss the limitations of existing methods and demonstrate the advantages of their model over seven other methods through extensive benchmarking studies on both simulated data under various scenarios and real datasets across different scales. This model achieves leading performance on both batch correction and biological information conservation and maintains scalable to datasets with over half a million cells.
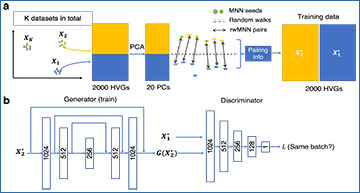
Workflow of ResPAN
(a) Generation of training data based on random walk mutual nearest neighbors (rwMNN) pairs. Pairs are generated in the top 20 PC space, but the training data are in the gene space using all cells contained in the rwMNN pairs. (b) Training process. We utilize the adversarial training strategy to optimize our model. The generator is a fully skip connected version of residual autoencoder. (c) Data integration and visualization. The results received from the generator are utilized for downstream analysis. Notations: X1: the selected reference batch; X2: the first query batch; 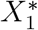 : the reference training data;
: the reference training data;  : the query training data; G: the generator.
: the query training data; G: the generator.
Availability – An open-source implementation of ResPAN and scripts to reproduce the results can be downloaded from: https://github.com/AprilYuge/ResPAN.
Wang Y, Liu T, Zhao H. (2022) ResPAN: a powerful batch correction model for scRNA-seq data through residual adversarial networks. Bioinformatics [Epub ahead of print]. [abstract]