Tumors are complex masses composed of malignant and non-malignant cells. Variation in tumor purity (proportion of cancer cells in a sample) can both confound integrative analysis and enable studies of tumor heterogeneity. Researchers at A*STAR have developed PUREE, which uses a weakly supervised learning approach to infer tumor purity from a tumor gene expression profile. PUREE was trained on gene expression data and genomic consensus purity estimates from 7864 solid tumor samples. PUREE predicted purity with high accuracy across distinct solid tumor types and generalized to tumor samples from unseen tumor types and cohorts. Gene features of PUREE were further validated using single-cell RNA-seq data from distinct tumor types. In a comprehensive benchmark, PUREE outperformed existing transcriptome-based purity estimation approaches. Overall, PUREE is a highly accurate and versatile method for estimating tumor purity and interrogating tumor heterogeneity from bulk tumor gene expression data, which can complement genomics-based approaches or be used in settings where genomic data is unavailable.
Overview of PUREE
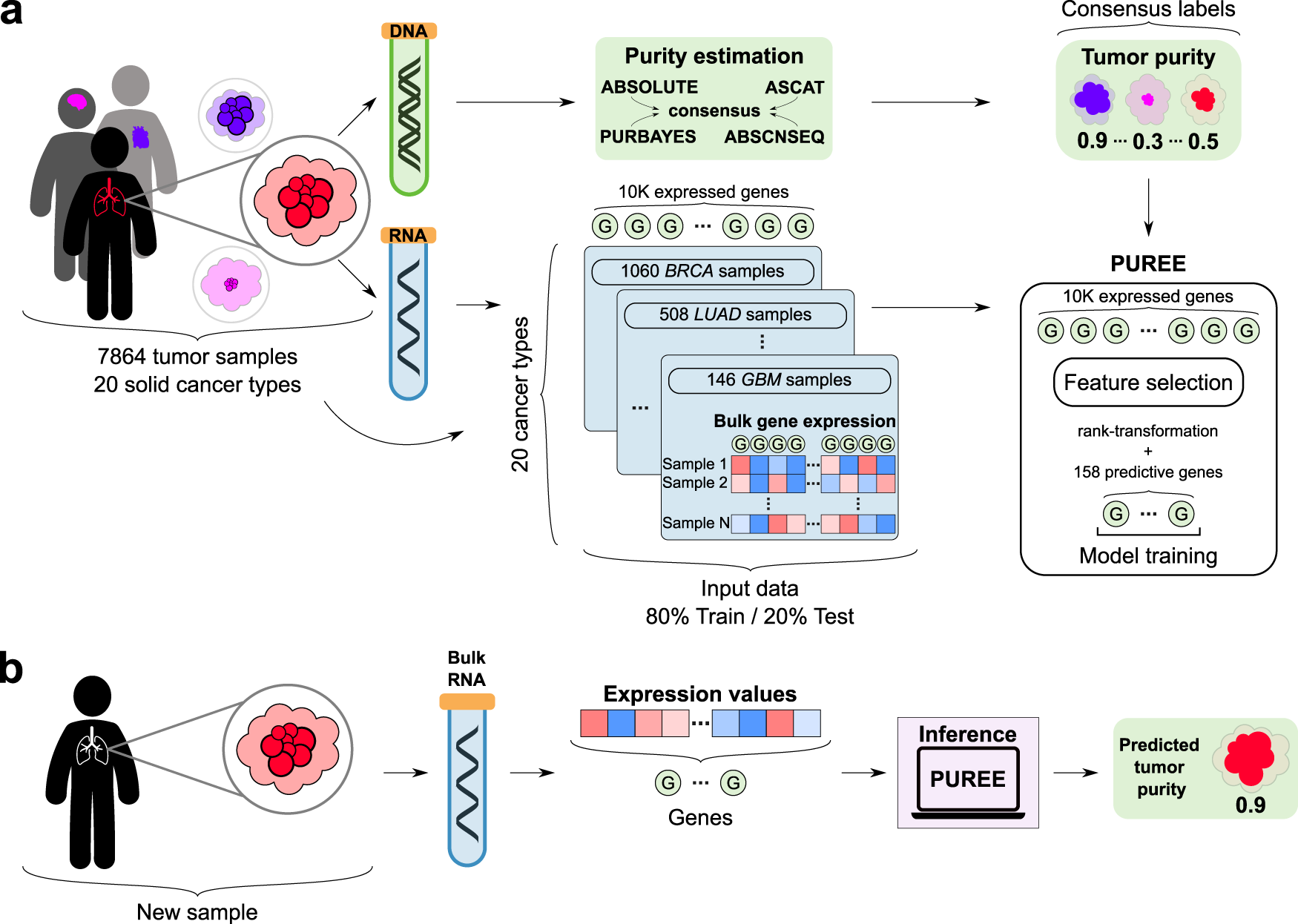
a PUREE is trained using a weakly supervised learning approach. Consensus genomics-based purity estimates are used as orthogonal (pseudo-ground-truth) labels, and a predictive model is trained on rank-transformed gene expression profiles from 7864 tumor samples spanning 20 solid tumor types (80%/20% train/test split). b For a new solid tumor sample, PUREE infers the purity from the corresponding tumor gene expression profile.
Availability – PUREE is available as a web service (https://puree.genome.sg/) and the respective Python package (https://github.com/skandlab/PUREE).
Revkov E, Kulshrestha T, Sung KW, Skanderup AJ. (2023) PUREE: accurate pan-cancer tumor purity estimation from gene expression data. Commun Biol 6(1):394. [article]