RNA Polymerase II (Pol II) is a multi-subunit complex that undergoes covalent modifications as transcription proceeds through genes and enhancers. Rate-limiting steps of transcription control Pol II recruitment, site and degree of initiation, pausing duration, productive elongation, nascent transcript processing, transcription termination, and Pol II recycling. Researchers at the KTH Royal Institute of Technology have developed Precision Run-On coupled to Immuno-Precipitation sequencing (PRO-IP-seq), which double-selects nascent RNAs and transcription complexes, and track phosphorylation of Pol II C-terminal domain (CTD) at nucleotide-resolution. The researchers uncover precise positional control of Pol II CTD phosphorylation as transcription proceeds from the initiating nucleotide (+1 nt), through early (+18 to +30 nt) and late (+31 to +60 nt) promoter-proximal pause, and into productive elongation. Pol II CTD is predominantly unphosphorylated from initiation until the early pause-region, whereas serine-2- and serine-5-phosphorylations are preferentially deposited in the later pause-region. Upon pause-release, serine-7-phosphorylation rapidly increases and dominates over the region where Pol II assembles elongation factors and accelerates to its full elongational speed. Interestingly, tracking CTD modifications upon heat-induced transcriptional reprogramming demonstrates that Pol II with phosphorylated CTD remains paused on thousands of heat-repressed genes. These results uncover dynamic Pol II regulation at rate-limiting steps of transcription and provide a nucleotide-resolution technique for tracking composition of engaged transcription complexes.
PRO-IP-seq maps molecular modifications of nascent transcription complexes
at nucleotide resolution
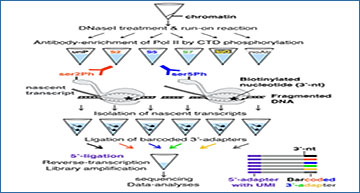
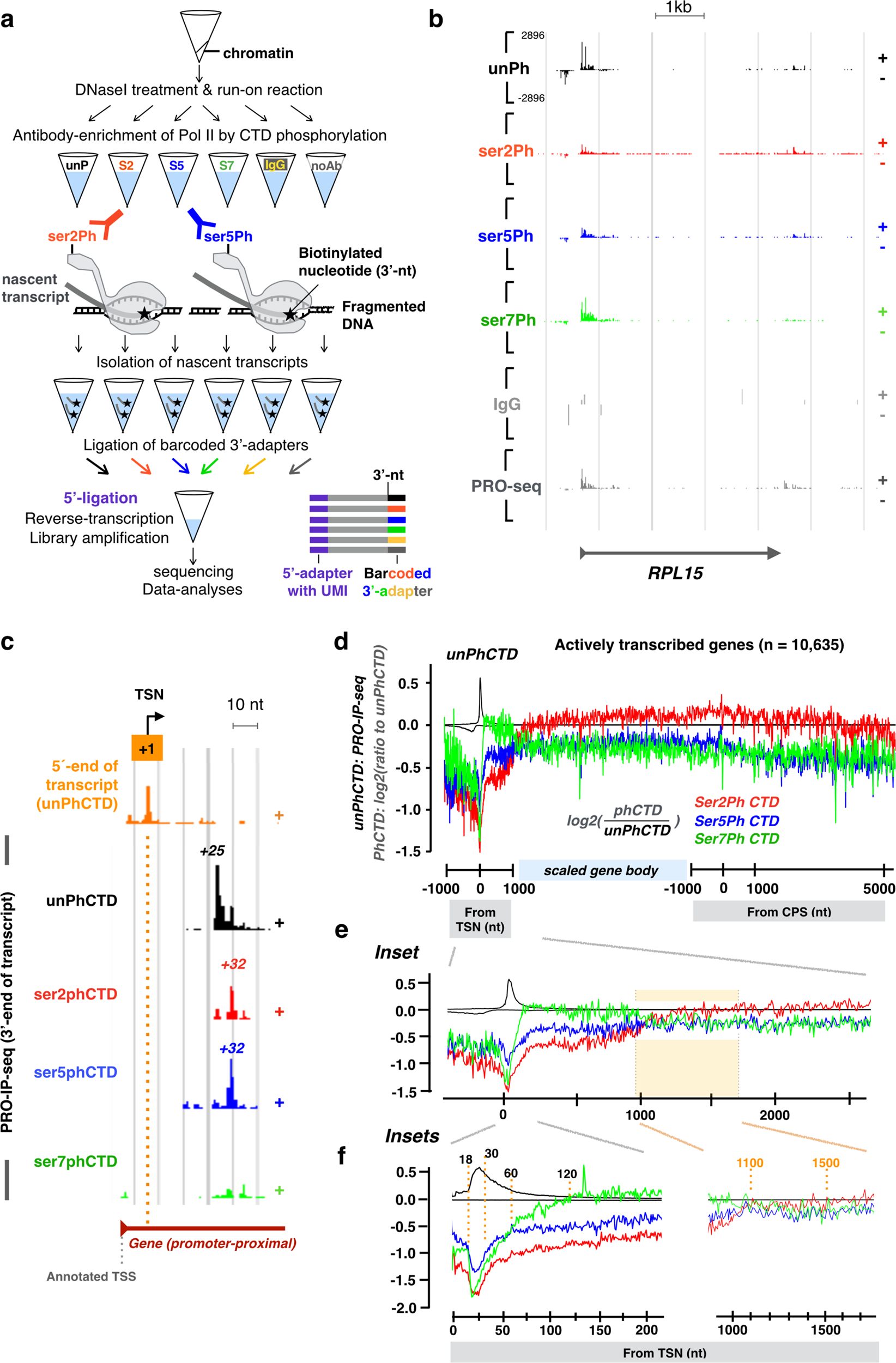
a Schematic presentation of the biochemical steps of PRO-IP-seq: Isolated chromatin is fragmented and a single biotinylated nucleotide incorporated into active sites of transcription. Transcription complexes are selected with antibodies against the C-terminal domain of Pol II (CTD), and nascent transcripts separated from non-nascent. The nascent transcripts are ligated to barcoded 3’-adapters, and the barcoded samples pooled for equal handling. After removal of 5’-cap, UMI-containing 5’-adapters are ligated. S2, S5 and S7 refer to phosphorylations of serine residues at Pol II CTD. b Representative example gene, RPL15, showing distribution of engaged Pol II with indicated status of CTD in non-treated K562 cells. The Y-scale is linear and the same in all tracks. c 5’-nt of PRO-IP-selected nascent RNAs enrich at the precise transcription start nucleotide (TSN; +1 nt), whereas the 3’-nt indicates the site of active transcription. The position of the highest Pol II pause coordinate from the +1 nt is indicated for each Pol II CTD modification. d Comparison of distinct CTD phosphorylations relative to the unphosphorylated form of Pol II CTD across the promoter-proximal region (linear scale from −1000 to +1000 nt from the TSN, +1 nt), gene body (scaled to 50 bins per gene), and CPS & termination window (linear scale from −1000 to +5000 from the CPS). The CTD modifications were mapped across all actively transcribed genes in untreated human K562 cells (n = 10,635). Average PRO-IP-signal is indicated for unphosphorylated Pol II CTD (black transcription profile), and the ratio of phosphorylated relative to unphophorylated CTD (red, green and blue) shown in log2 scale. e, f Insets of the promoter-proximal and early gene body regions. The dotted orange lines indicate nt-resolution distance from the TSN. In (e–f), each window is 1 nt. The Y-scales are as in d.
Availability – Pipelines for mapping the raw data and generating density profiles are available in GitHub (https://github.com/Vihervaara/PRO-IP-seq).
Vihervaara A, Versluis P, Himanen SV, Lis JT. (2023) PRO-IP-seq tracks molecular modifications of engaged Pol II complexes at nucleotide resolution. Nat Commun 14(1):7039. [article]