Single-cell RNA sequencing (scRNA-seq) is widely used to reveal heterogeneity in cells, which has given us insights into cell–cell communication, cell differentiation, and differential gene expression. However, analyzing scRNA-seq data is a challenge due to sparsity and the large number of genes involved. Therefore, dimensionality reduction and feature selection are important for removing spurious signals and enhancing the downstream analysis.
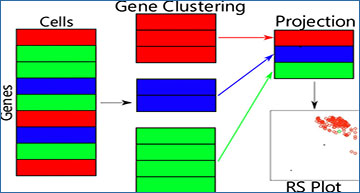
Researchers at Michigan State University have developed Correlated Clustering and Projection (CCP), a new data-domain dimensionality reduction method, for the first time. CCP projects each cluster of similar genes into a supergene defined as the accumulated pairwise nonlinear gene–gene correlations among all cells. Using 14 benchmark data sets, the researchers demonstrate that CCP has significant advantages over classical principal component analysis (PCA) for clustering and/or classification problems with intrinsically high dimensionality. In addition, they introduce the Residue-Similarity index (RSI) as a novel metric for clustering and classification and the R-S plot as a new visualization tool. The researchers show that the RSI correlates with accuracy without requiring the knowledge of the true labels. The R-S plot provides a unique alternative to the uniform manifold approximation and projection (UMAP) and t-distributed stochastic neighbor embedding (t-SNE) for data with a large number of cell types.
Hozumi Y, Tanemura KA, Wei GW. (2023) Preprocessing of Single Cell RNA Sequencing Data Using Correlated Clustering and Projection. J Chem Inf Model [Epub ahead of print]. [abstract]