Targeted RNA sequencing (RNA-seq) from FFPE specimens is used clinically in cancer for its ability to estimate gene expression and to detect fusions. Using a cohort of NSCLC patients, researchers at the Peter MacCallum Cancer Centre sought to determine whether targeted RNA-seq could be used to measure tumour mutational burden (TMB) and the expression of immune-cell-restricted genes from FFPE specimens and whether these could predict response to immune checkpoint blockade.
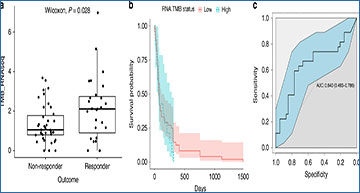
Using The Cancer Genome Atlas LUAD dataset, the researchers developed a method for determining TMB from tumour-only RNA-seq and showed a correlation with DNA sequencing derived TMB calculated from tumour/normal sample pairs (Spearman correlation = 0.79, 95% CI [0.73, 0.83]. They applied this method to targeted sequencing data from our patient cohort and validated these results against TMB estimates obtained using an orthogonal assay (Spearman correlation = 0.49, 95% CI [0.24, 0.68]). The researchers observed that the RNA measure of TMB was significantly higher in responders to immune blockade treatment (P = 0.028) and that it was predictive of response (AUC = 0.640 with 95% CI [0.493, 0.786]). By contrast, the expression of immune-cell-restricted genes was uncorrelated with patient outcome.
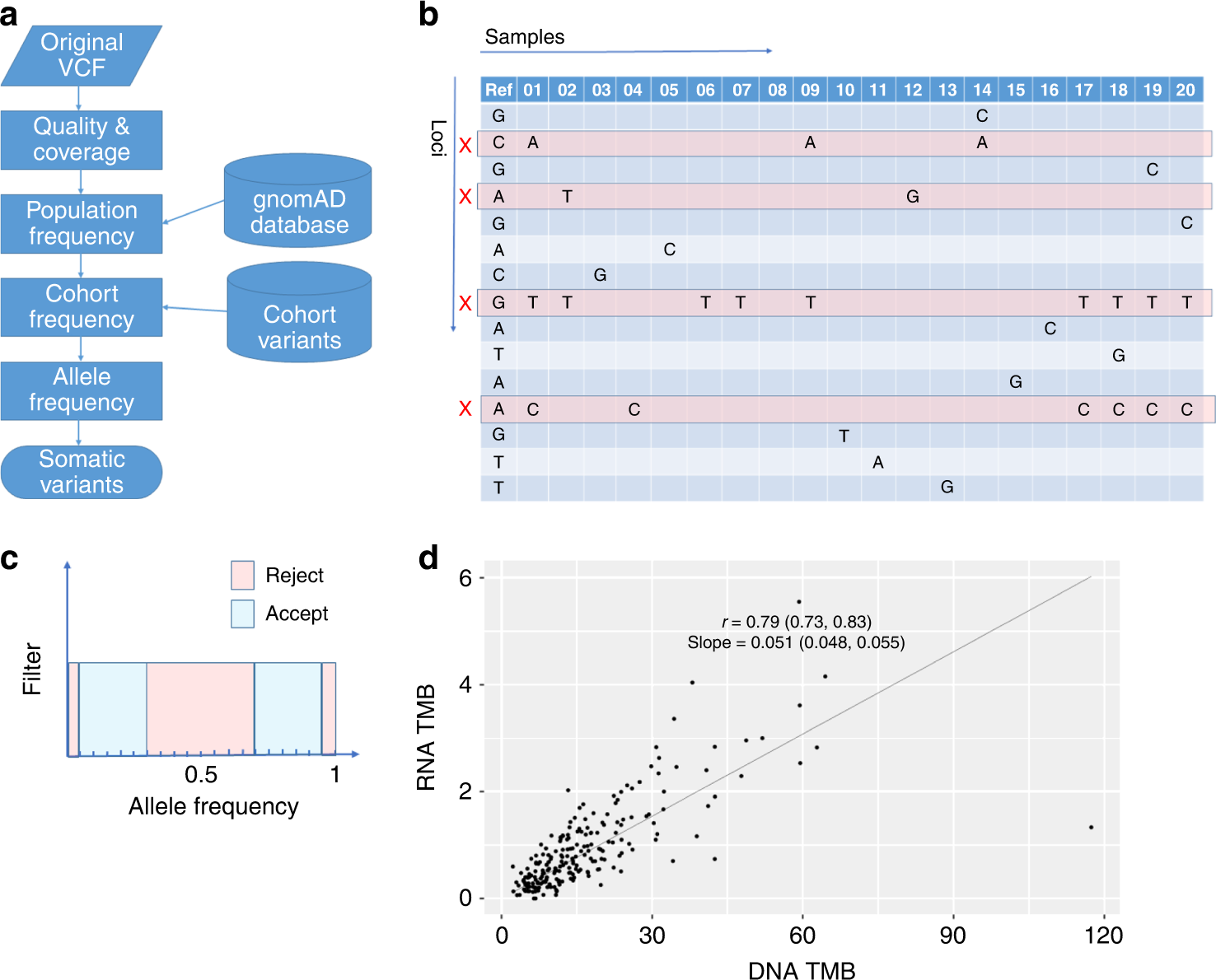
Filtering variants from tumour-only RNA-seq to compute RNA TMB
a Flowchart illustrating the sequence of filtering steps used to remove unwanted variants. b All variants in the cohort are aggregated into a table containing loci as rows and samples as columns. This enables the identification of loci-containing multiple variants. These commonly occurring variants (with loci marked by a red cross in the figure) are unlikely to be due to randomly distributed mutations and hence can be removed from TMB calculations based on their frequency within the cohort. c Variants are accepted or rejected based on their allele frequency in each sample. VAFs close to 0 or 1 are removed as are those in a range around 0.5. d RNA TMB versus DNA TMB for the TCGA-LUAD dataset with a linear regression fitted through the origin.
Markham JF, Fellowes AP, Green T, Leal JL, Legaie R, Cullerne D, Morris T, John T, Solomon B, Fox SB. (2023) Predicting response to immune checkpoint blockade in NSCLC with tumour-only RNA-seq. Br J Cancer [Epub ahead of print]. [abstract]