With the upcoming of affordable Next-Generation Sequencing technologies, the number of known non-protein coding RNAs increased drastically in recent years. Different types of non-coding RNAs (ncRNAs) emerged as key players in the regulation of gene expression on the RNA-RNA, RNA-DNA as well as RNA-protein level, ranging from involvement in chromatin remodeling and transcription regulation to post-transcriptional modifications. Prediction of ncRNAs involves the use of several bioinformatics tools and can be a daunting task for researchers. This led to the development of analysis pipelines such as UClncR and lncpipe. However, these pipelines are limited to datasets from human, mouse, zebrafish or fruit fly and are not able to analyze RNA sequencing data from other organisms.
Researchers at the Vienna University of Technology have developed the analysis pipeline Pinc (Pipeline for prediction of ncRNA) as an enhanced tool to predict ncRNAs based on sequencing data by removing transcripts that show protein-coding potential. Additionally, a feature for differential expression analysis of annotated genes as well as for identification of novel ncRNAs is implemented. Pinc uses Nextflow as a framework and is built with robust and well-established analysis tools. This will allow researchers to utilize sequencing data from every organism in order to reliably identify ncRNAs.
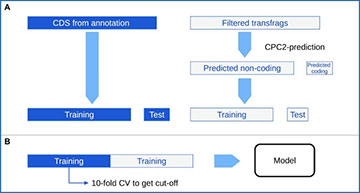
Graphical overview on Pinc
Raw Sequencing reads are filtered based on quality and length using fastp. Subsequently, HISAT2 aligns the reads against the reference genome. StringTie assembles aligned reads into transfrags. Transfrags of already annotated features are removed by filtering for putative novel ncRNAs based on gffcompare’s transfrag classification code. Together with the protein-coding RNAs from the reference annotation an organism-specific model is trained using CPC2 and CPAT to assess the coding probability of all putative, novel, non-coding transfrags. As edgeR requires the total count of reads mapped to each transfrag for a differential expression analysis, HTSeq-count was used to count the reads.
Availability – Pinc is accessible as a Nextflow pipeline on GitHub: https://github.com/brummetheus/pinc
Schmal M, Girod C, Yaver D, Mach RL, Mach-Aigner AR. (2022) A bioinformatic-assisted workflow for genome-wide identification of ncRNAs. NAR Genom Bioinform 4(3):lqac059. [article]