Splicing (represented by the scissors) in noncoding parts of mRNA enhances its export from the nucleus to the cytoplasm, which promotes oncogene expression in tumor cells.
An interactive web portal developed by scientists at KAUST offers a platform for cancer researchers to interrogate how RNA splicing in noncoding parts of genes fuels the growth of different types of tumors.
Using splice junctions specifically located within 3′ UTRs, the scientists identified and quantified 3USPs in 7,917 RNA sequencing (RNA-seq) samples across ten cancer types from The Cancer Genome Atlas (TCGA) and the corresponding tissues from The Genotype-Tissue Expression (GTEx)
The new resource, named SpUR (short for Splicing in Untranslated Regions) and freely available online, details more than 1,000 splicing events found frequently in cancers in noncoding regions of mRNA located just downstream of protein-coding stop signals. The sites and expression levels of these events are catalogued and visualized for nearly 8,000 samples across 10 cancer types and corresponding normal tissues.
With the tool, independent research team can now further probe the role of individual splice events in cancer development and progression.
“These events could become candidates to study RNA dysregulations in cancer for academic researchers,” says Xin Gao, acting associate director of the Computational Bioscience Research Center and deputy director of the Smart Health Initiative at KAUST. “Or they could serve as a primary source for the development of RNA-based anti-cancer drugs.”
Computer scientist Gao, together with postdoc Bin Zhang and research engineer Adil Salhi, created the SpUR database in collaboration with researchers at the Cancer Science Institute of Singapore.
The research showed that splicing in downstream sequences of a gene (known as 3’ untranslated regions, or 3’ UTRs) is pervasive in cancers, especially in genes linked to tumor aggression. Consequently, patients whose cancers harbor more of these gene-restructuring events tend to have poorer survival outcomes.
As a proof of principle, the researchers designed splice-switching agents known as antisense oligonucleotides (ASOs) that could block this splicing process in 3’ UTRs. When administered to liver cancer cells, these drugs helped repress tumor growth.
And since the same kinds of splicing events are “ubiquitously expressed across different cancer types,” Gao notes, this type of therapeutic strategy “could be helpful to develop broad-spectrum anti-cancer drugs.”
Upregulated 3′ UTR splicing may promote carcinogenesis
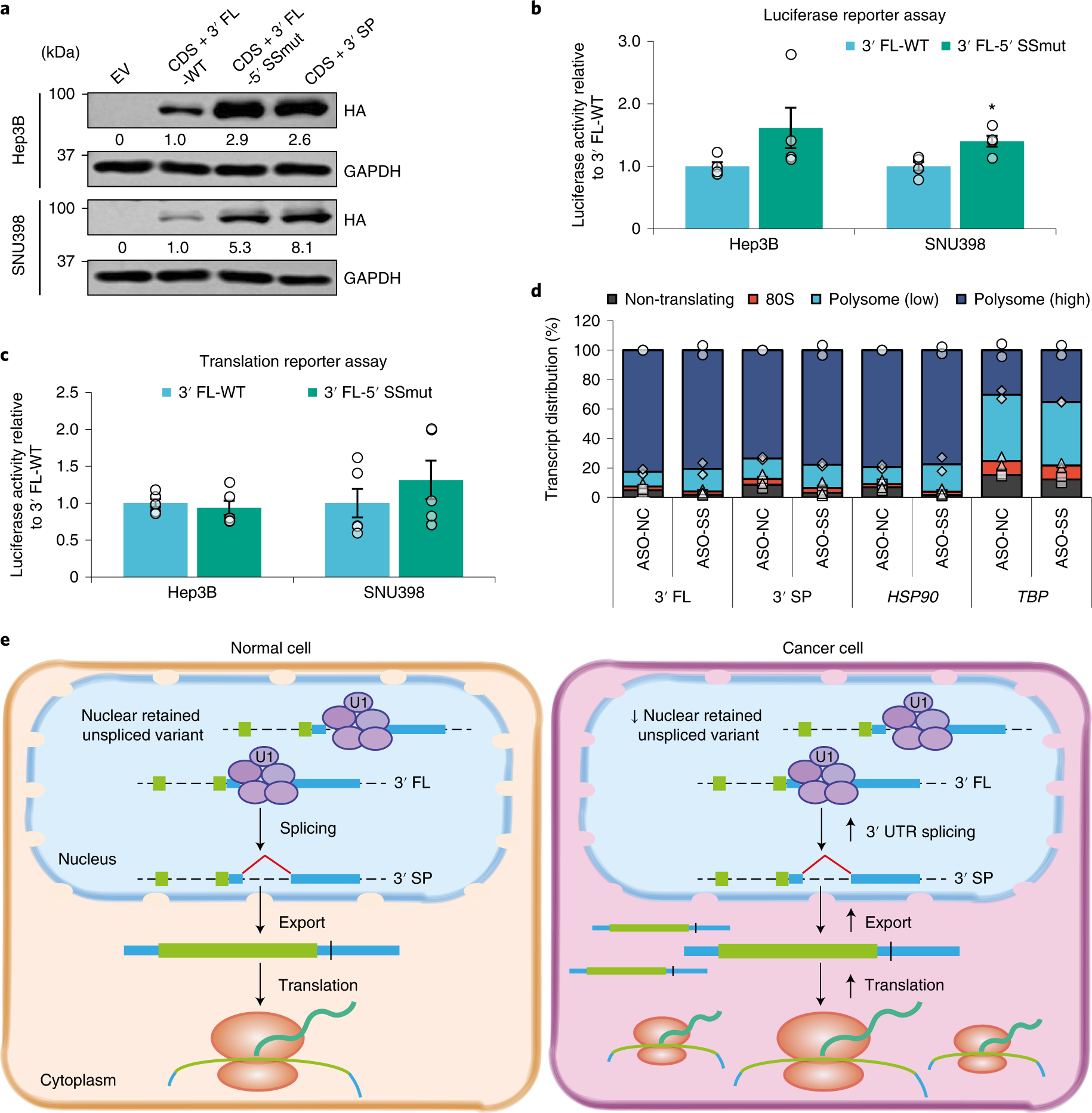
a, Effect of overexpressing CTNNB1 CDS + 3′ FL-WT, CDS + 3′ FL-5′ SSmut and CDS + 3′ SP on exogenous CTNNB1 protein expression. Data represent three independent experiments. b,c, Luciferase activity of plasmid-transfected (n = 4 independent experiments) (b) and RNA-transfected (n = 5 independent experiments) (c) CTNNB1 3′ FL-WT and 3′ FL-5′ SSmut reporter constructs. d, Effect of ASO-mediated blocking of the CTNNB1 3′ UTR splice site on the polysome profiles for CTNNB1 (n = 2 independent experiments). HSP90 and TBP are housekeeping controls. e, Schematic depicting how increased 3′ UTR splicing may promote carcinogenesis. In b and c: mean ± s.e.m.; unpaired Student’s t-test *P < 0.05, **P < 0.01 and ***P < 0.001.
One potential target: CTNNB1, which is a gene that provides instructions for making a protein called beta-catenin. Drug companies have long tried to target beta-catenin, given its central role in many cancer-signaling pathways — but with only limited success. The study from Gao and his collaborators showed that splicing in the 3’ UTR of CTNNB1 is widespread across cancers of the liver, breast, colon, kidney, lung and other organs, and that a spliced variant is the predominant driver of tumor progression.
In a mouse model of liver cancer, blocking this splicing resulted in complete tumor regression. An ASO therapy directed at CTNNB1 splicing could therefore have broad utility in patients, and, as Gao points out, it’s not likely to be the only one.
Source – King Abdullah University of Science & Technology (KAUST)
Availability – https://www.cbrc.kaust.edu.sa/spur/home/
Chan JJ, Zhang B, Chew XH, Salhi A, Kwok ZH, Lim CY, Desi N, Subramaniam N, Siemens A, Kinanti T, Ong S, Sanchez-Mejias A, Ly PT, An O, Sundar R, Fan X, Wang S, Siew BE, Lee KC, Chong CS, Lieske B, Cheong WK, Goh Y, Fam WN, Ooi MG, Koh BTH, Iyer SG, Ling WH, Chen J, Yoong BK, Chanwat R, Bonney GK, Goh BKP, Zhai W, Fullwood MJ, Wang W, Tan KK, Chng WJ, Dan YY, Pitt JJ, Roca X, Guccione E, Vardy LA, Chen L, Gao X, Chow PKH, Yang H, Tay Y. (2022) Pan-cancer pervasive upregulation of 3′ UTR splicing drives tumourigenesis. Nat Cell Biol [Epub ahead of print]. [article]