The human genome is a vast landscape of genetic information, but much of it remains shrouded in mystery. Up to 80% of the genome produces what scientists call “dark matter” RNAs—mysterious molecules that do not code for proteins. These noncoding RNAs (ncRNAs) play crucial roles in regulating gene expression, but their full diversity and functions have remained elusive. However, recent advancements in sequencing technology have allowed researchers to shed light on this hidden world of noncoding RNA.
A groundbreaking study by researchers at Sun Yat-sen University introduces a novel method called NAP-seq, which enables scientists to globally profile noncapped RNAs (napRNAs) with various terminal modifications at single-nucleotide resolution. Using this innovative approach, the researchers uncovered a vast array of structured ncRNAs that were previously unknown. These include stably expressed linear intron RNAs (sliRNAs), snoRNA-intron RNAs (snotrons), RNAs embedded in miRNA spacers (misRNAs), and thousands of other structured napRNAs in both humans and mice.
NAP-seq accurately identifies full-length napRNAs at single-nucleotide resolution
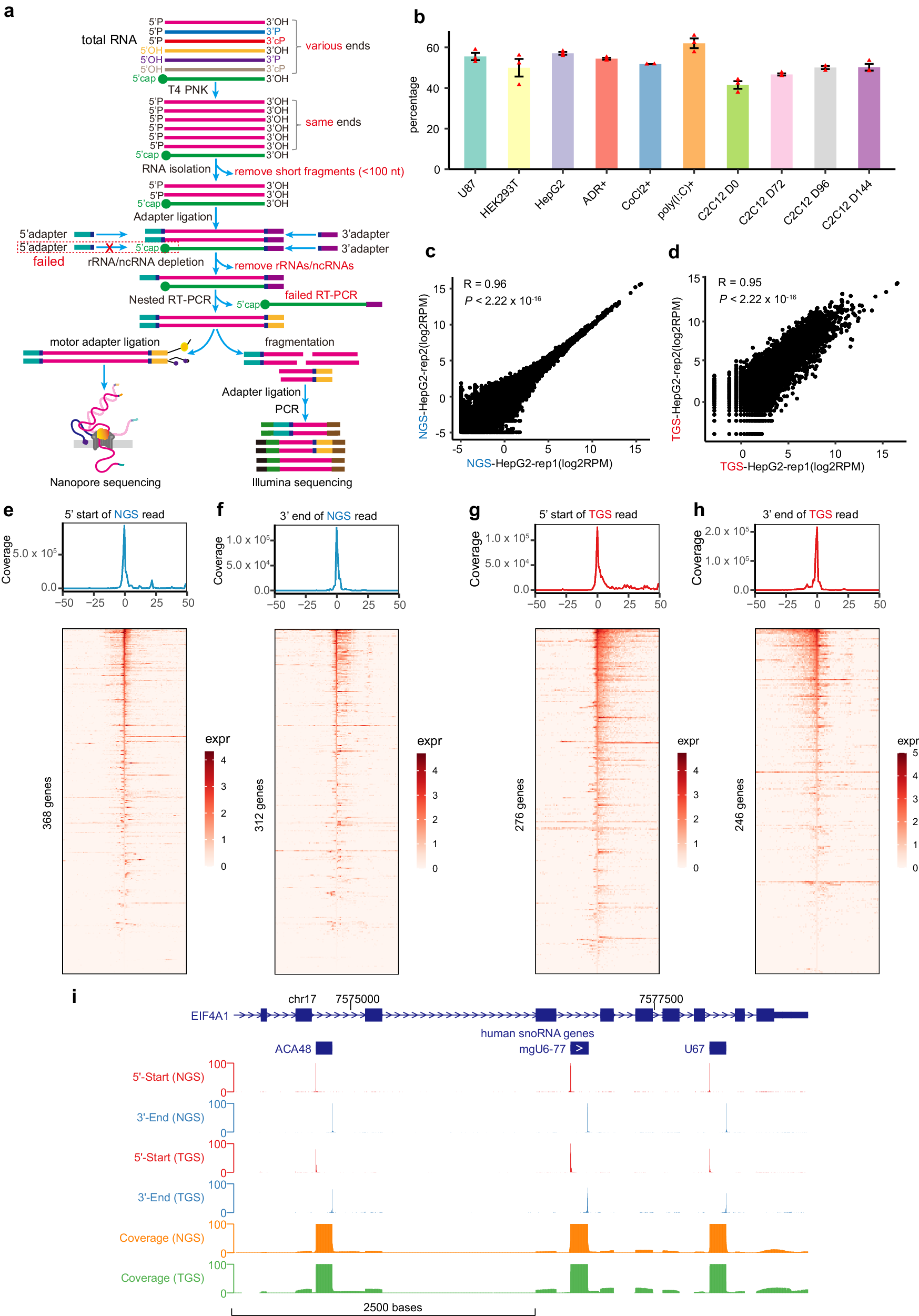
a Schematic depiction of the NAP-seq method with the construction of TGS libraries and NGS libraries. Total RNA was extracted and pre-size selection of RNA fractions was performed by removing short RNAs. b The percentage of reads with at least one specific adapter in the NAP-seq-NGS libraries in humans and mice (3 biological replicates per sample). All data are plotted as the means ± SEMs. The number of reads (RPM, reads per million) for known ncRNAs was highly reproducible between replicates with both the NAP-seq-NGS (c) and NAP-seq-TGS (d) methods. p values were calculated by two-sided Pearson’s correlation test. e, f Coincidence analysis of both terminal sites in the NAP-seq-NGS reads and those in the known ncRNAs in HepG2 cells. The x-axis shows the distance from the annotated 5’-start (e) or 3’-end (f) site in the NAP-seq-NGS read to the corresponding terminal sites in the known ncRNAs, and the y-axis shows the number of reads (RPM) within a certain distance from the 5’-start or 3’-end site. The bottom panel shows a heatmap, in which each row represents a gene that matches the terminal sites with the NAP-seq-NGS read exactly, and each column represents the expression values of genes at a specific distance. expr, expression value. g Number of coincidences between the 5’-start sites in NAP-seq-TGS reads and the 5’-start sites in known ncRNAs in HepG2 cells. h Number of coincidences between the 3’-end sites in NAP-seq-TGS reads and the 3’-end sites in known ncRNAs in HepG2 cells. i Genome Browser visualization of NAP-seq 5’-start, 3’-end and coverage signals (RPM, reads per million), which were generated from NAP-seq-NGS and NAP-seq-TGS data, in a 5000-bp region of a human snoRNA cluster (chr17:7,573,750-7,578,750).
The study revealed that these structured ncRNAs undergo dynamic changes in response to different stimuli and during cellular differentiation. Importantly, the researchers identified specific napRNAs that play critical roles in cellular processes. For example, one structured napRNA was found to regulate myoblast differentiation—a key step in muscle development. Another napRNA, known as DINAP, was shown to interact with dyskerin pseudouridine synthase 1 (DKC1) to promote cell proliferation by maintaining DKC1 protein stability.
These findings have profound implications for our understanding of gene regulation and cellular function. By uncovering a diverse range of structured ncRNAs and elucidating their roles in various biological processes, this study opens new avenues for research into the functions of dark matter RNAs. Moreover, the identification of specific napRNAs involved in key cellular processes suggests potential targets for therapeutic intervention in diseases where these processes are dysregulated.
The discovery of structured ncRNAs through NAP-seq represents a significant advancement in our understanding of the noncoding universe. By revealing the complexity and functional diversity of dark matter RNAs, this study paves the way for future research aimed at deciphering the intricate regulatory networks that govern gene expression and cellular behavior. Ultimately, unraveling the mysteries of dark matter RNAs holds the promise of unlocking new insights into human health and disease.
Liu S, Huang J, Zhou J, Chen S, Zheng W, Liu C, Lin Q, Zhang P, Wu D, He S, Ye J, Liu S, Zhou K, Li B, Qu L, Yang J. (2024) NAP-seq reveals multiple classes of structured noncoding RNAs with regulatory functions. Nat Commun 15(1):2425. [article]