Gene expression is characterised by stochastic bursts of transcription that occur at brief and random periods of promoter activity. The kinetics of gene expression burstiness differs across the genome and is dependent on the promoter sequence, among other factors. Single-cell RNA sequencing (scRNA-seq) has made it possible to quantify the cell-to-cell variability in transcription at a global genome-wide level. However, scRNA-seq data is prone to technical variability, including low and variable capture efficiency of transcripts from individual cells.
Researchers at Imperial College London have developed a novel mathematical theory for the observed variability in scRNA-seq data. Their method captures burst kinetics and variability in both the cell size and capture efficiency, which allows them to propose several likelihood-based and simulation-based methods for the inference of burst kinetics from scRNA-seq data. Using both synthetic and real data, the researchers show that the simulation-based methods provide an accurate, robust and flexible tool for inferring burst kinetics from scRNA-seq data. In particular, in a supervised manner, a simulation-based inference method based on neural networks proves to be accurate and useful when applied to both allele and non-allele-specific scRNA-seq data.
Model of stochastic gene expression and the effect of cell size and sequencing
capture efficiency on observed transcript count distributions
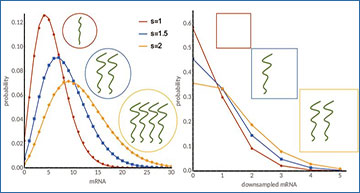
(a) An illustration of the telegraph model of stochastic gene expression and its associated parameters. Gene switches between an inactive and active state and mRNAs are transcribed only from the active state. (b) Illustration of downsampling in scRNA-seq with a constant β = 0.5 (note that in reality β tends to be smaller and varies across the cells. Effective transcription rate  is proportional to cell size in original transcript counts (right) and both cell size and capture efficiency in the observed counts (right). (c) Distributions of original mRNA counts in cells with constant size for three specific parameters sets for the telegraph model (left) and their corresponding downsampled distribution (right). Distribution of cell specific capture efficiencies (β) used in downsampling is illustrated in the middle top arrow. The challenge is to use the downsampled observed count distribution that is also affected by variability in capture efficiency and cell size to infer the parameters of the original distribution (middle bottom arrow).
is proportional to cell size in original transcript counts (right) and both cell size and capture efficiency in the observed counts (right). (c) Distributions of original mRNA counts in cells with constant size for three specific parameters sets for the telegraph model (left) and their corresponding downsampled distribution (right). Distribution of cell specific capture efficiencies (β) used in downsampling is illustrated in the middle top arrow. The challenge is to use the downsampled observed count distribution that is also affected by variability in capture efficiency and cell size to infer the parameters of the original distribution (middle bottom arrow).
Availability: The code for Neural Network and Approximate Bayesian Computation inference is available at https://github.com/WT215/nnRNA and https://github.com/WT215/Julia_ABC respectively.
Tang W, Jørgensen ACS, Marguerat S, Thomas P, Shahrezaei V. (2023) Modelling capture efficiency of single-cell RNA-sequencing data improves inference of transcriptome-wide burst kinetics. Bioinformatics [Epub ahead of print]. [abstract]