Single-cell RNA-seq (scRNA-seq) analysis of multiple samples separately can be costly and lead to batch effects. Exogenous barcodes or genome-wide RNA mutations can be used to demultiplex pooled scRNA-seq data, but they are experimentally or computationally challenging and limited in scope. Mitochondrial genomes are small but diverse, providing concise genotype information. Researchers at Shanghai Jiao Tong University have developed “mitoSplitter,” an algorithm that demultiplexes samples using mitochondrial RNA (mtRNA) variants, and demonstrated that mtRNA variants can be used to demultiplex large-scale scRNA-seq data. Using affordable computational resources, mitoSplitter can accurately analyze 10 samples and 60,000 cells in 6 h. To avoid the batch effects from separated experiments, the researchers applied mitoSplitter to analyze the responses of five non-small cell lung cancer cell lines to BET (Bromodomain and extraterminal) chemical degradation in a multiplexed fashion. They found the synthetic lethality of TOP2A inhibition and BET chemical degradation in BET inhibitor-resistant cells. The result indicates that mitoSplitter can accelerate the application of scRNA-seq assays in biomedical research.
Mitochondrial variants as the optimal material for genotyping
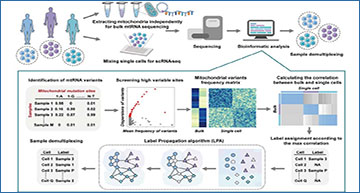
(A) The mean Spearman correlation between all pairs of two individuals in a published RNA sequencing data based on mitochondrial or autosomal variant profiles. The red line represents the mean correlation of the mitochondrial variants between individuals, while the upper and lower pink boundaries represent the first and third quantiles. MT represents “mitochondrion”. These results demonstrated that mitochondrial variant profiles are significantly more variable between individuals than those of other chromosomes. (B) The mean Spearman correlation between pairwise individuals from a published RNA sequencing data based on variant profiles of mitochondrial or autosomal segments. An autosomal segment is composed of 13 adjacently located genes (equal to the gene number of the mitochondrial genome), and 100 segments were randomly extracted from each autosomal segment. The red line represents the mean correlation of the mitochondrial genome between individuals. MT represents “mitochondrion”. These results indicate that profiles of mitochondrial variants are significantly more variable than those of other chromosome segments containing the same number of genes. (C) Overview of mitoSplitter framework. SNPs were identified by aligning RNA-seq data to the reference genome for each sample. High-variability mitochondrial SNPs were identified based on variation frequency and dispersions in bulk RNA-seq, and used to genotype samples. The mitochondrial genotyping correlation between bulk samples and cells was used to determine the sample label of some single cells. Only cells with high bulk sample correction were labeled. Then, the label propagation algorithm (LPA) was employed to give sample labels to unlabeled cells based on single-cell mtRNA SNPs.
Availability – The code is available on https://github.com/lnscan/mitoSplitter.
Lin X, Chen Y, Lin L, Yin K, Cheng R, Lin X, Wang X, Guo Y, Wu Z, Zhang Y, Li J, Yang C, Song J. (2023) mitoSplitter: A mitochondrial variants-based method for efficient demultiplexing of pooled single-cell RNA-seq. PNAS 120(39):e2307722120. [abstract]