The joint analysis of the genome, epigenome, transcriptome, proteome and/or metabolome from single cells is transforming our understanding of cell biology in health and disease. In less than a decade, the field has seen tremendous technological revolutions that enable crucial new insights into the interplay between intracellular and intercellular molecular mechanisms that govern development, physiology and pathogenesis. Researchers from the University of Leuven highlight advances in the fast-developing field of single-cell and spatial multi-omics technologies (also known as multimodal omics approaches), and the computational strategies needed to integrate information across these molecular layers. They demonstrate their impact on fundamental cell biology and translational research, discuss current challenges and provide an outlook to the future.
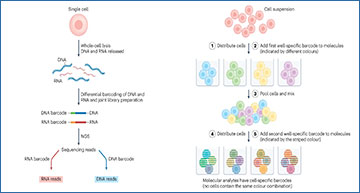
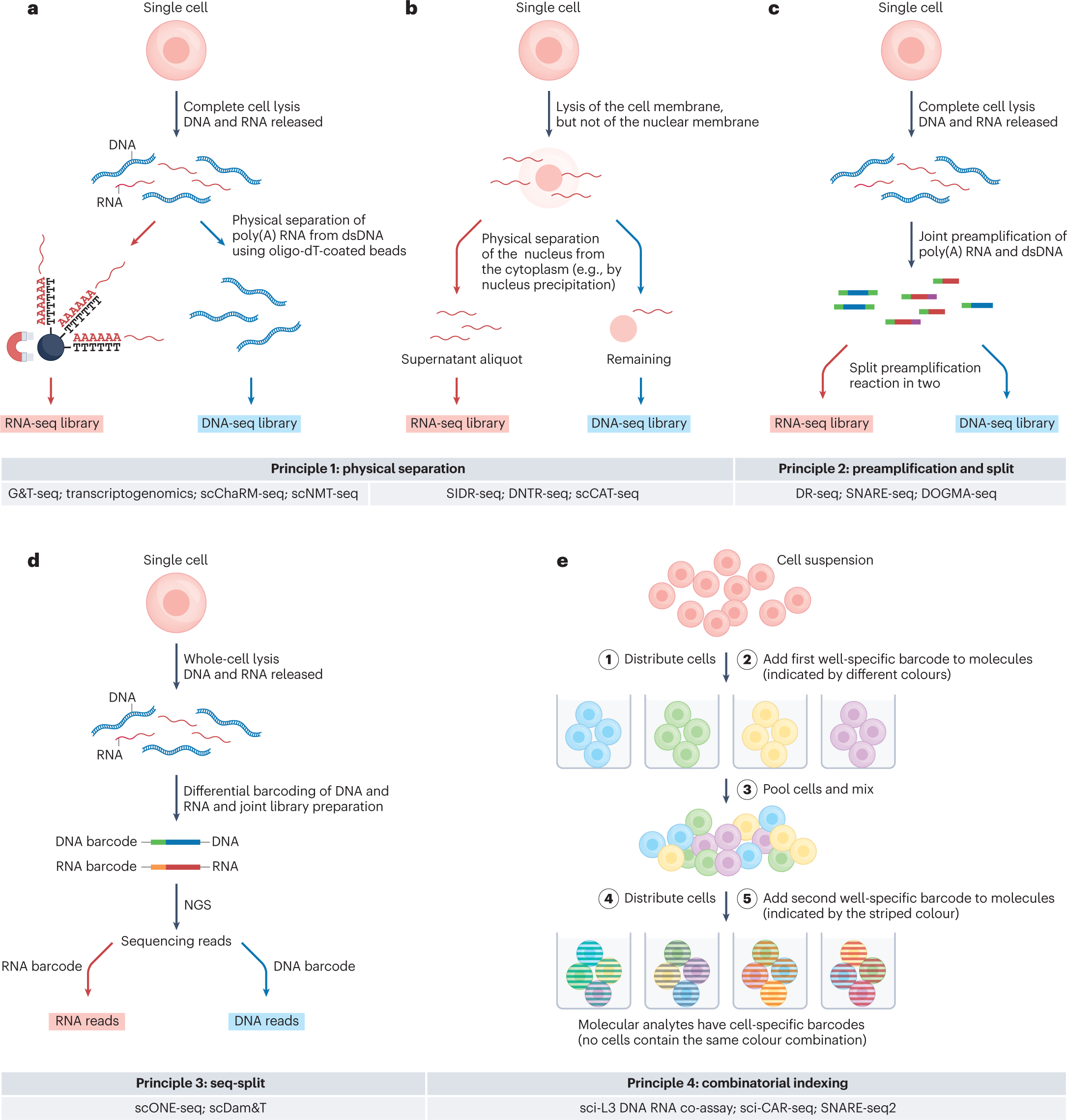
The four general principles for multi-ome measurements from single cells
All principles are visualized with RNA and DNA as example analytes. Principle 1 is based on physical separation of the distinct molecular analytes (parts a,b). a, Following complete lysis of the isolated cell or nucleus, poly(A) RNA hybridizes to oligo-dT-coated paramagnetic beads, and following magnetic pulldown, the supernatant that contains the genomic DNA is transferred to a new reaction vessel b, In an alternative approach, lysis conditions that rupture the cell but not the nuclear envelope allow separation of nuclear from cytoplasmic molecular analytes, either by precipitating the nucleus with centrifugation or magnetic pulldown followed by aspiration of the cytosolic supernatant or by microfluidic-controlled nucleus-from-cytoplasm separation. c, In principle 2, termed preamplification and split, distinct analytes are differentially tagged and jointly preamplified, followed by splitting the preamplification reaction for analyte-specific sequencing library preparations. d, Principle 3, termed seq-split, involves analyte-specific barcoding and sequencing library preparation in a single-pot reaction. e, In principle 4, termed combinatorial indexing, molecular analytes of single cells are tagged without isolating single cells.
Vandereyken K, Sifrim A, Thienpont B, Voet T. (2023) Methods and applications for single-cell and spatial multi-omics. Nat Rev Genet [Epub ahead of print]. [article]