Long-read RNA sequencing has arisen as a counterpart to short-read sequencing, with the potential to capture full-length isoforms, albeit at the cost of lower depth. Yet this potential is not fully realized due to inherent limitations of current long-read assembly methods and underdeveloped approaches to integrate short-read data. Researchers from the University of Chicago, critically compare the existing methods and develop a new integrative approach to characterize a particularly challenging pool of low-abundance long noncoding RNA (lncRNA) transcripts from short- and long-read sequencing in two distinct cell lines. Their analysis reveals severe limitations in each of the sequencing platforms. For short-read assemblies, coverage declines at transcript termini resulting in ambiguous ends, and uneven low-coverage results in segmentation of a single transcript into multiple transcripts. Conversely, long-read transcript assembly lacks strand-of-origin information and depth, culminating in erroneous assembly and quantitation of transcripts. The researchers also discover a cDNA synthesis artifact in long-read datasets that markedly impacts the identity and quantitation of assembled transcripts. Towards remediating these problems, they develop a computational pipeline to ″strand″ long-read cDNA libraries that rectifies inaccurate mapping and assembly of long-read transcripts. Leveraging the strengths of each platform and our computational stranding, they present and benchmark a hybrid assembly approach that drastically increases the sensitivity and accuracy of full-length transcript assembly on the correct strand and improves detection of biological features of the transcriptome. When applied to a challenging set of under-annotated and cell-type variable lncRNA, our method resolves the segmentation problem of short-read sequencing and the depth problem of long-read sequencing, resulting in the assembly of coherent transcripts with precise 5′ and 3′ ends. This workflow can be applied to existing datasets for superior demarcation of transcript ends and refined isoform structure, which can enable better differential gene expression analyses and molecular manipulations of transcripts.
Improved transcript assembly with strand-aware hybrid pipeline
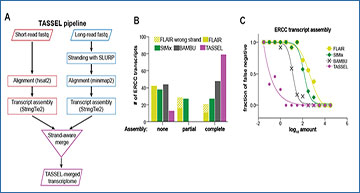
A. Workflow of TASSEL (Transcript Assembly using Short and Strand-Emended Long reads) pipeline. Transcripts obtained from short-read RNA-seq are merged with those obtained from stranded long reads in a strand-aware manner. B. Number and extent of ERCC standard transcripts assembled by the indicated assembly method in the HAP1 dataset. StMix, StringTie Mix. C. False negative rate of assembling ERCC transcripts, by each of the indicated assembly methods, as a function of their abundance in the HAP1 dataset. D. Ends of the 92 ERCC transcripts (arranged in the increasing order of length) assembled by TASSEL (magenta circle) and StringTie Mix (StMix, green diamond) in the HAP1 dataset. Gray bar indicates actual transcript. The color bar indicates the abundance of the given transcript. E. Sensitivity of the indicated assembly methods at the locus level. Transcriptome assembled by the given method in the HAP1 dataset was compared against reference annotation (gencode hg38v35) using gffcompare. F. Percent of assem bled transcripts that match completely with a transcript (left) or contained within an intron (right) of reference transcript (gencode hg38v35), using StringTie Mix or TASSEL in the HAP1 dataset. G. Proximity of TSS (left) and TTS (right) of known genes (gencode hg38v41)to the TSS and TTS of transcripts assembled by TASSEL or StringTie Mix in the HAP1 dataset. H. Enrichment of H3K4me3 (left, normalized to input) and RNA Pol II (right, normalized to the total number of mapped reads) at the TSS of transcripts assembled by TASSEL or StringTie Mix in the HAP1 dataset. H3K4me3 and RNA Pol II occupancy calculated from ChIP-seq data from HAP1 cells. I. Average PRO-seq signal at TSS of transcripts assembled by TASSEL or StringTie Mix on the positive (top) and negative (bottom) strands.
Kainth AS, Haddad GA, Hall JM, Ruthenburg AJ. (2022) Merging short and stranded long reads improves transcript assembly. bioRXiv [online preprint]. [abstract]