Deep learning methods are widely applied in digital pathology to address clinical challenges such as prognosis and diagnosis. As one of the most recent applications, deep models have also been used to extract molecular features from whole slide images. Although molecular tests carry rich information, they are often expensive, time-consuming, and require additional tissue to sample.
Researchers at the Mayo Clinic have developed tRNAsformer, an attention-based topology that can learn both to predict the bulk RNA-seq from an image and represent the whole slide image of a glass slide simultaneously. The tRNAsformer uses multiple instance learning to solve a weakly supervised problem while the pixel-level annotation is not available for an image. The researchers conducted several experiments and achieved better performance and faster convergence in comparison to the state-of-the-art algorithms. The proposed tRNAsformer can assist as a computational pathology tool to facilitate a new generation of search and classification methods by combining the tissue morphology and the molecular fingerprint of the biopsy samples.
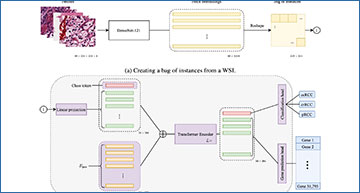
A diagram showing how tRNAsformer works
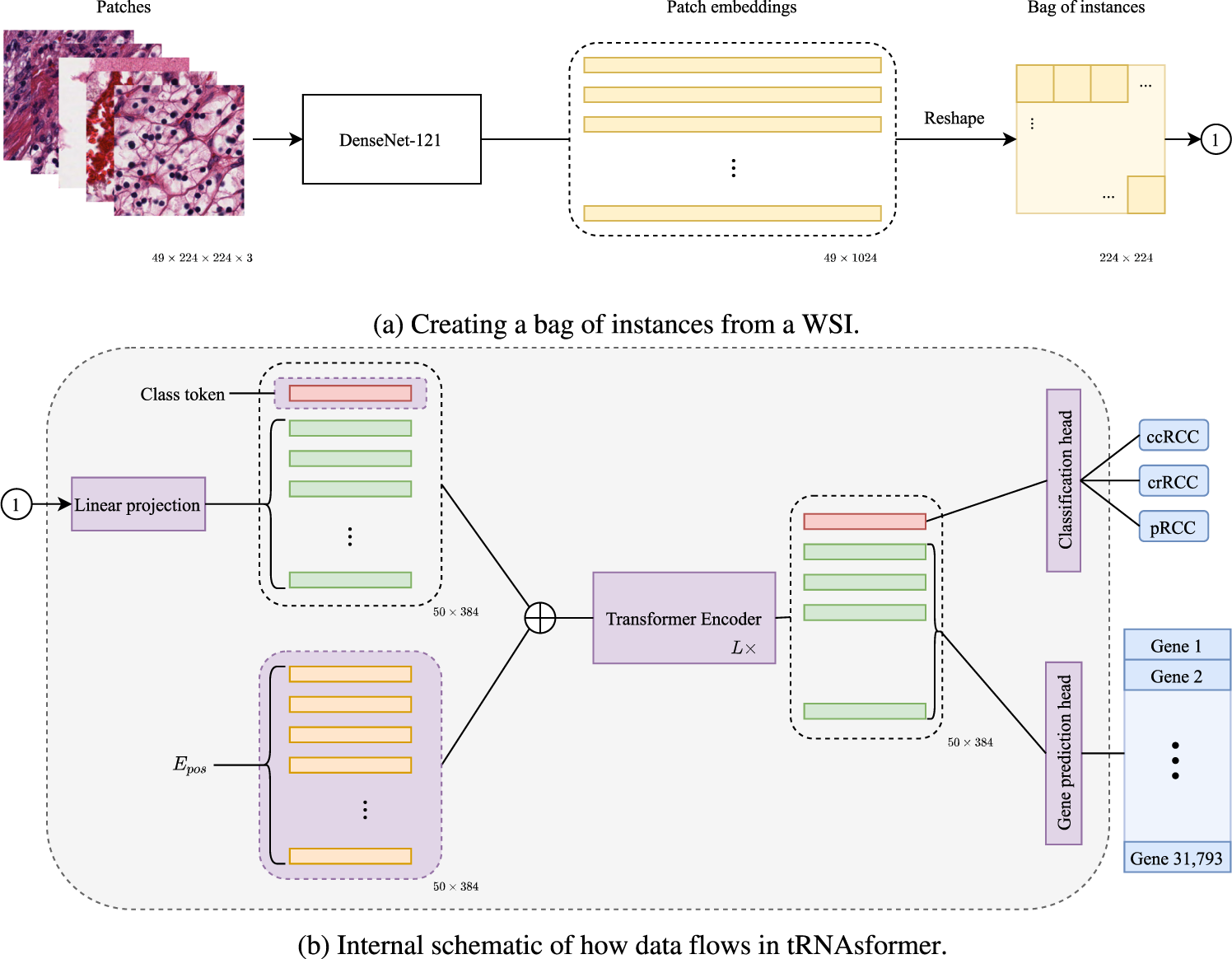
a 49 tiles of size 224 × 224 × 3 selected from 49 spatial clusters in a WSI are embedded with a DenseNet-121. The outcome is a matrix of size 49 × 1024 as DenseNet-121 has 1024 deep features after the last pooling. Then the matrix is reshaped and rearranged to 224 × 224 matrix in which each 32 × 32 block corresponds to a tile embedding 1 × 1024. b Applying a 2D convolution with kernel 32, stride 32, and 384 kernels, each 32 × 32 block has linearly mapped a vector of 384 dimensional. Next, a class token is concatenated with the rest of the tile embeddings, and Epos is added to the matrix before entering L Encoder layers. The first row of the outcome, which is associated with the class token, is fed to the classification head. The rest of the internal embeddings that are associated with all tile embeddings are passed to the gene prediction head. All parts with learnable variables are shown in purple.
Availability – the source code along with the trained tRNAsformer models are available at https://doi.org/10.5281/zenodo.7613349.
Alsaafin A, Safarpoor A, Sikaroudi M, Hipp JD, Tizhoosh HR. (2023) Learning to predict RNA sequence expressions from whole slide images with applications for search and classification. Commun Biol 6(1):304. [article]