RNA fate and function are affected by their structures and interactomes. However, how RNA and RNA-binding proteins (RBPs) assemble into higher-order structures and how RNA molecules may interact with each other to facilitate functions remain largely unknown. Researchers at the University of Chicago have developed KARR-seq, which uses N3-kethoxal labeling and multifunctional chemical crosslinkers to covalently trap and determine RNA–RNA interactions and higher-order RNA structures inside cells, independent of local protein binding to RNA. KARR-seq depicts higher-order RNA structure and detects widespread intermolecular RNA–RNA interactions with high sensitivity and accuracy. Using KARR-seq, the researchers show that translation represses mRNA compaction under native and stress conditions. They determined the higher-order RNA structures of respiratory syncytial virus (RSV) and vesicular stomatitis virus (VSV) and identified RNA–RNA interactions between the viruses and the host RNAs that potentially regulate viral replication.
KARR-seq maps higher-order RNA structures
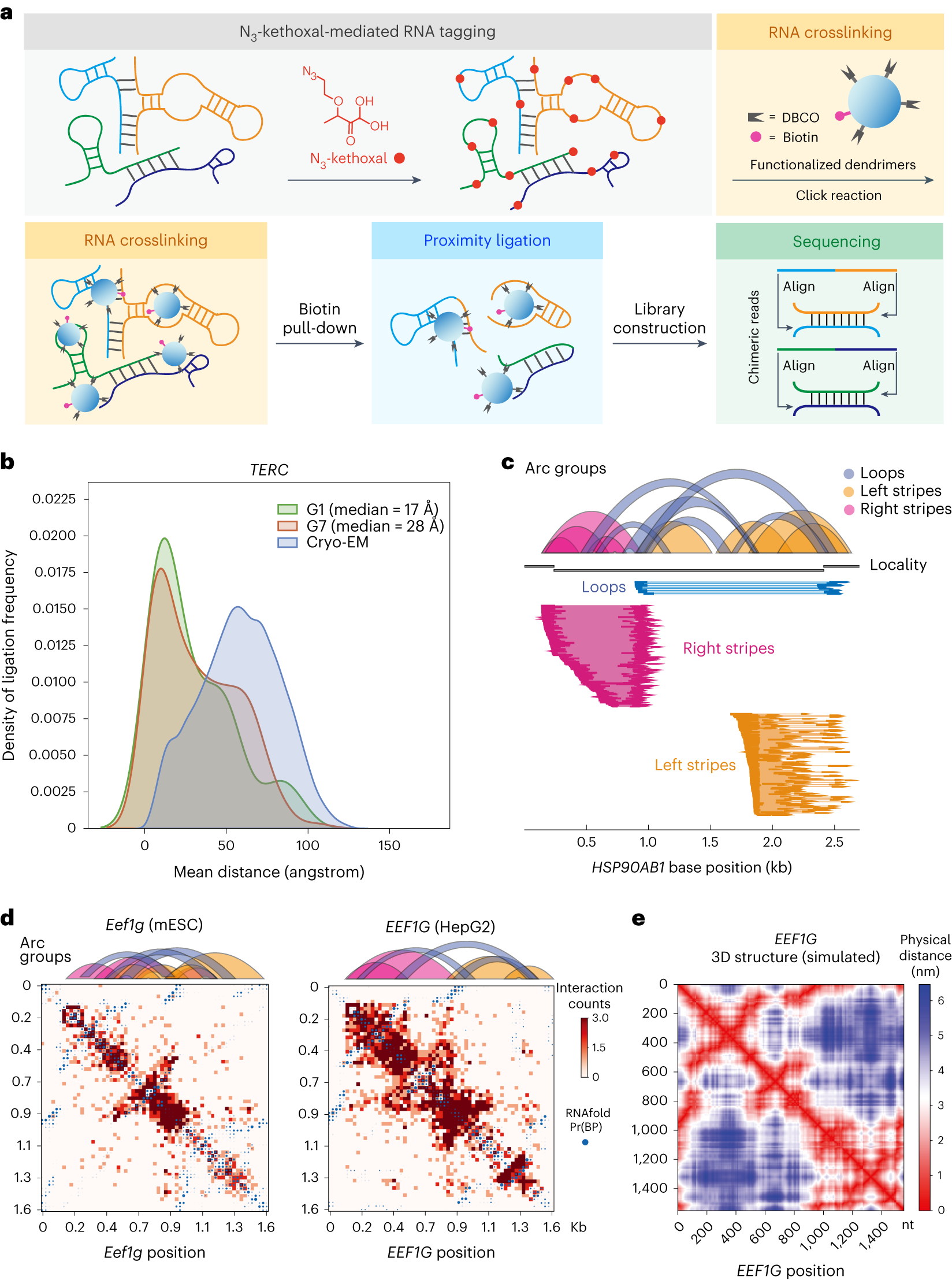
a, KARR-seq workflow. Cells are first treated with N3-kethoxal to modify RNAs with azide tags (red), which enables crosslinking of the tagged RNA molecules by DBCO-decorated dendrimers (blue). Biotin modifications (pink) on dendrimers facilitate the enrichment of crosslinking products, followed by proximity ligation, RNA library construction and sequencing. Chimeric sequencing reads are aligned to identify RNA–RNA interactions. b, Physical distances between interacting fragments of TERC in K562 cells, measured by KARR-seq data generated using G1 and G7 dendrimers, respectively. The physical distances were measured using the cryo-EM structure of TERC. The actual physical distance distribution in the cryo-EM structure is shown in blue for comparison. c, Illustration of loop and stripe structures detected by KARR-seq. In arc groups, loops, left stripes and right stripes are denoted in blue, yellow and pink, respectively. Corresponding KARR-seq chimeric reads are displayed below. d, The KARR-seq interaction maps and arc groups for the Eef1g (EEF1G) transcript in mESCs (left) and HepG2 cells (right). e, The simulated physical distance map of the human EEF1G transcript. For b–d, KARR-seq was performed in two biological replicates.
Availability – The source code of KARR-seq is freely available at https://github.com/ouyang-lab/KARR-seq
Wu T, Cheng AY, Zhang Y et al. (2023) KARR-seq reveals cellular higher-order RNA structures and RNA–RNA interactions. Nat Biotechnol [Epub ahead of print]. [article]