Bulk RNA-sequencing technologies have provided invaluable insights into host and bacterial gene expression and associated regulatory networks. Nevertheless, the majority of these approaches report average expression across cell populations, hiding the true underlying expression patterns that are often heterogeneous in nature. Due to technical advances, single-cell transcriptomics in bacteria has recently become reality, allowing exploration of these heterogeneous populations, which are often the result of environmental changes and stressors.
Researchers from the University of Würzburg have improved their previously published bacterial single-cell RNA-sequencing protocol that is based on MATQ-seq, achieving a higher throughput through the integration of automation. The researchers also selected a more efficient reverse transcriptase, which led to reduced cell loss and higher workflow robustness. Moreover, they successfully implemented a Cas9-based ribosomal RNA depletion protocol into the MATQ-seq workflow. Applying their improved protocol on a large set of single Salmonella cells sampled over growth revealed improved gene coverage and a higher gene detection limit compared to their original protocol and allowed us to detect the expression of small regulatory RNAs, such as GcvB or CsrB at a single-cell level. In addition, the researchers confirmed previously described phenotypic heterogeneity in Salmonella in regards to expression of pathogenicity-associated genes. Overall, the low percentage of cell loss and high gene detection limit makes the improved MATQ-seq protocol particularly well suited for studies with limited input material, such as analysis of small bacterial populations in host niches or intracellular bacteria.
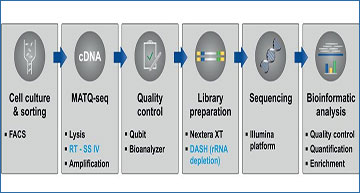
Improved MATQ-seq workflow for bacterial single-cell RNA-seq
A) Overview of bacterial scRNA-seq pipeline including major steps from cell culture to bioinformatic analysis. Changes compared to the previous MATQ-seq protocol are highlighted in blue. B) Detailed workflow of the MATQ-seq protocol separated into two main steps: (left) cell isolation and cDNA synthesis; (right) library preparation including DASH for ribosomal RNA depletion. Major improvements are highlighted in blue, including the use of SuperScript IV for reverse transcription, reaction optimization and integration of DASH into the library preparation. All pipetting steps were automated using the I.DOT dispensing robot, with the exception of all clean-up and quality control steps.
Homberger C, Hayward RJ, Barquist L, Vogel J. (2022) Improved bacterial single-cell RNA-seq through automated MATQ-seq and Cas9-based removal of rRNA reads. bioRXiv [online preprint]. [article]