There is a strong need to eliminate batch-specific differences when integrating single-cell RNA-sequencing (scRNA-seq) datasets generated under different experimental conditions for downstream task analysis. Existing batch correction methods usually transform different batches of cells into one preselected “anchor” batch or a low-dimensional embedding space, and cannot take full advantage of useful information from multiple sources.
Researchers from the China University of Petroleum have developed a novel framework, called IMGG, i.e., integrating multiple single-cell datasets through connected graphs and generative adversarial networks (GAN) to eliminate nonbiological differences between different batches. Compared with current methods, IMGG shows excellent performance on a variety of evaluation metrics, and the IMGG-corrected gene expression data incorporate features from multiple batches, allowing for downstream tasks such as differential gene expression analysis.
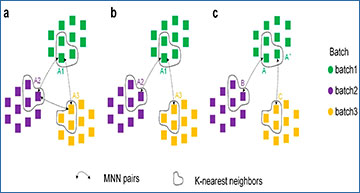
Overview of the IMGG framework
(a) Data preprocessing. (b) In the embedding space, three methods are used to construct connected graphs. (c) Combining the connected graphs and HVGs, sampling from similar cells, and using GAN to correct batch effects.
Wang X, Zhang C, Zhang Y, Meng X, Zhang Z, Shi X, Song T. (2022) IMGG: Integrating Multiple Single-Cell Datasets through Connected Graphs and Generative Adversarial Networks. Int J Mol Sci 23(4):2082. [article]