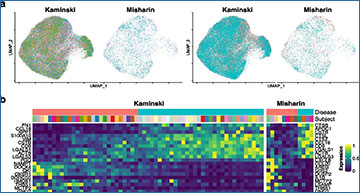
Single-cell RNA sequencing (scRNA-seq) technology has enabled assessment of transcriptome-wide changes at single-cell resolution. Due to the heterogeneity in environmental exposure and genetic background across subjects, subject effect contributes to the major source of variation in scRNA-seq data with multiple subjects, which severely confounds cell type specific differential expression (DE) analysis. Moreover, dropout events are prevalent in scRNA-seq data, leading to excessive number of zeroes in the data, which further aggravates the challenge in DE analysis.
Researchers at the Yale School of Public Health have developed iDESC to detect cell type specific DE genes between two groups of subjects in scRNA-seq data. iDESC uses a zero-inflated negative binomial mixed model to consider both subject effect and dropouts. The prevalence of dropout events (dropout rate) was demonstrated to be dependent on gene expression level, which is modeled by pooling information across genes. Subject effect is modeled as a random effect in the log-mean of the negative binomial component. The researchers evaluated and compared the performance of iDESC with eleven existing DE analysis methods. Using simulated data, they demonstrated that iDESC had well-controlled type I error and higher power compared to the existing methods. Applications of those methods with well-controlled type I error to three real scRNA-seq datasets from the same tissue and disease showed that the results of iDESC achieved the best consistency between datasets and the best disease relevance.
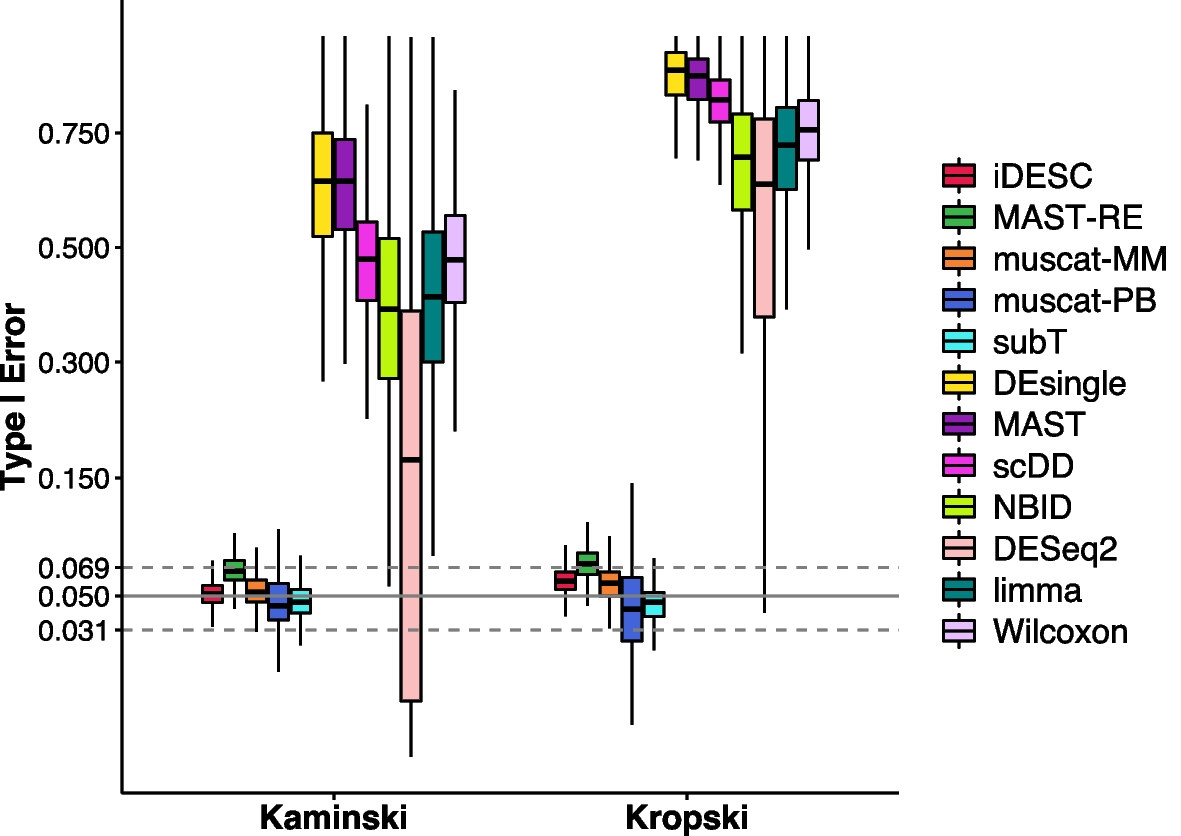
Empirical type I error of all methods on the two permuted real datasets
Boxplots showing the median (center line), interquartile range (hinges), and 1.5 times the interquartile (whiskers) of empirical type I error at the nominal level of 0.05. Confidence interval of type I error is marked by two dashed lines (0.031–0.069)
iDESC was able to achieve more accurate and robust DE analysis results by separating subject effect from disease effect with consideration of dropouts to identify DE genes, suggesting the importance of considering subject effect and dropouts in the DE analysis of scRNA-seq data with multiple subjects.
Availability – An R package implementing the proposed method is available at https://github.com/yl883/iDESC under the MIT license and was deposited to Zenodo (https://doi.org/10.5281/zenodo.6929851).
Liu Y, Zhao J, Adams TS, Wang N, Schupp JC, Wu W, McDonough JE, Chupp GL, Kaminski N, Wang Z, Yan X. (2023) iDESC: identifying differential expression in single-cell RNA sequencing data with multiple subjects. BMC Bioinformatics 24(1):318. [article]