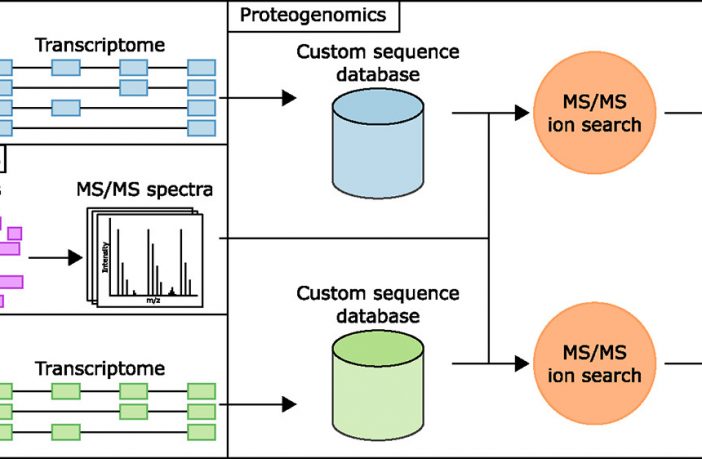
Alternative splicing can lead to distinct protein isoforms. These can have different functions in specific cells and tissues or in different developmental stages. In this study, researchers from the University of New South Wales explored whether transcripts assembled from long read, nanopore-based, direct RNA-sequencing (RNA-seq) could improve the identification of protein isoforms in human K562 cells. By comparing with Illumina-based short read RNA-seq, the researchers showed that a large proportion of Ensembl transcripts (5949/14,326) and genes expressing alternatively spliced transcripts (486/2981) identified with long direct reads were missed by short paired-end reads. By co-analyzing proteomic and transcriptomic data, they also showed that some peptides (826/35,976), proteins (262/3215), and protein isoforms arising from distinct transcript variants (574/1212) identified with isoform-specific peptides via custom long-read-based databases were missed in Illumina-derived databases. Finally, we generated unequivocal peptide evidence for a set of protein isoforms and showed that long read, direct RNA-seq allows the discovery of novel protein isoforms not already in reference databases or custom databases built from short read RNA-seq data. Our analysis highlights the benefits of long read RNA-seq data in the generation of reference databases to increase tandem mass spectrometry (MS/MS) identification of protein isoforms.
Tay AP, Hamey JJ, Martyn GE, Wilson LOW, Wilkins MR. (2022) Identification of Protein Isoforms Using Reference Databases Built from Long and Short Read RNA-Sequencing. J Proteome Res [Epub ahead of print]. [article]