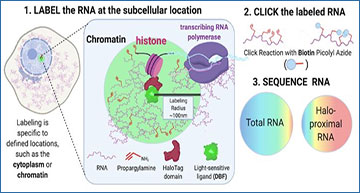
The subcellular localization of specific RNA molecules promotes localized cellular activity across a variety of species and cell types. The misregulation of this RNA targeting can result in developmental defects, and mutations in proteins that regulate this process are associated with multiple diseases. For the vast majority of localized RNAs, however, the mechanisms that underlie their subcellular targeting are unknown, partly due to the difficulty associated with profiling and quantifying subcellular RNA populations. To address this challenge, researchers at University of Colorado Anschutz Medical Campus have developed Halo-seq, a proximity labeling technique that can label and profile local RNA content at virtually any subcellular location. Halo-seq relies on a HaloTag fusion protein localized to a subcellular space of interest. Through the use of a radical-producing Halo ligand, RNAs that are near the HaloTag fusion are specifically labeled with spatial and temporal control. Labeled RNA is then specifically biotinylated in vitro via a click reaction, facilitating its purification from a bulk RNA sample using streptavidin beads. The content of the biotinylated RNA is then profiled using high-throughput sequencing. In this article, the researchers describe the experimental and computational procedures for Halo-seq, including important benchmark and quality control steps. By allowing the flexible profiling of a variety of subcellular RNA populations, they envision Halo-seq facilitating the discovery and further study of RNA localization regulatory mechanisms.
Lo HG, Engel KL, Goering R, Li Y, Spitale RC, Taliaferro JM. (2022) Halo-seq: An RNA Proximity Labeling Method for the Isolation and Analysis of Subcellular RNA Populations. Curr Protoc 2(5):e424. [abstract]