Recent developments in single-cell RNA sequencing have opened up a multitude of possibilities to study tissues at the level of cellular populations. However, the heterogeneity in single-cell sequencing data necessitates appropriate procedures to adjust for technological limitations and various sources of noise when integrating datasets from different studies. While many analysis procedures employ various preprocessing steps, they often overlook the importance of selecting and optimizing the employed data transformation methods.
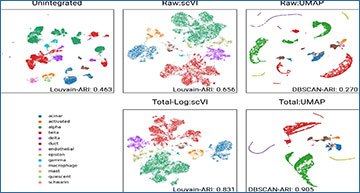
Researchers at the University Medical Center Göttingen investigated data transformation approaches used in single-cell clustering analysis tools and their effects on batch integration analysis. In particular, the researchers compared 16 transformations and their impact on the low-dimensional representations, aiming to reduce the batch effect and integrate multiple single-cell sequencing data. Their results show that data transformations strongly influence the results of single-cell clustering on low-dimensional data space, such as those generated by UMAP or PCA. Moreover, these changes in low-dimensional space significantly affect trajectory analysis using multiple datasets, as well. However, the performance of the data transformations greatly varies across datasets, and the optimal method was different for each dataset. Additionally, they explored how data transformation impacts the analysis of deep feature encodings using deep neural network-based models, including autoencoder-based models and proto-typical networks. Data transformation also strongly affects the outcome of deep neural network models.
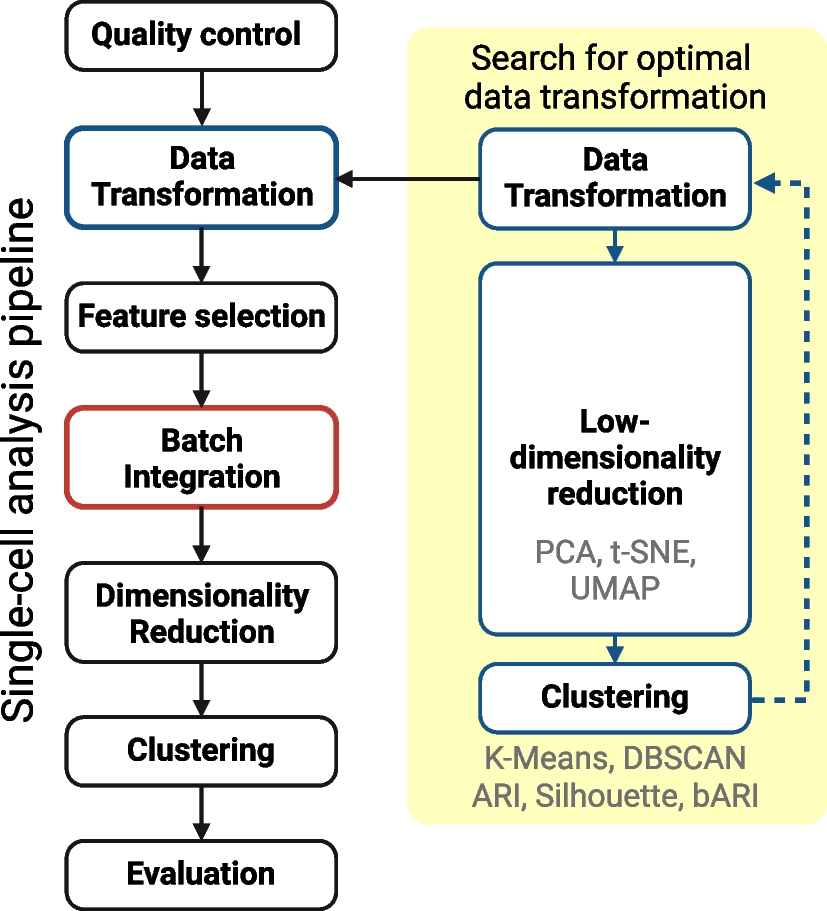
Overview of the proposed low-dimensional analysis workflow for conducting a thorough search for appropriate data transformations
The researchers evaluated the effect of data transformation while integrating different batches of single-cell RNA sequencing data. For that, they tested 16 different data transformations with subsequent dimensionality reduction methods and clustering algorithms and compared their results. This single-cell analysis in conventional practices has feature selection, batch integration, and dimensionality reduction.
These findings suggest that the batch effect and noise in integrative analysis are highly influenced by data transformation. Low-dimensional features can integrate different batches well when proper data transformation is applied. Furthermore, the researchers found that the batch mixing score on low-dimensional space can guide the selection of the optimal data transformation. In conclusion, data preprocessing is one of the most crucial analysis steps and needs to be cautiously considered in the integrative analysis of multiple scRNA-seq datasets.
Park Y, Hauschild AC. (2024) The effect of data transformation on low-dimensional integration of single-cell RNA-seq. BMC Bioinformatics 25(1):171. [article]