RNA sequencing has paved the way for transcriptomics, which is the study of all the molecules that interact with ribonucleic acid (RNA) in a cell. It has revolutionized the study of gene expression and has become a vital tool for understanding the molecular mechanisms underlying various diseases. A new study by researchers at the Karolinska Institutet researchers showcases the importance of RNA sequencing in investigating the effects of environmental exposures on human neural development.
The study used a fractional factorial experimental design-based transcriptomics (FFED) approach to investigate the effects of low-grade environmental exposures on differentiating human neural progenitors. The study utilized six environmental factors (lead, valproic acid, bisphenol A, ethanol, fluoxetine hydrochloride, and zinc deficiency) and four human induced pluripotent stem cell line-derived differentiating human neural progenitors to showcase the utility of the FFED coupled with RNA-sequencing in identifying the effects of these environmental factors. The study used a layered analytical approach and detected several convergent and divergent gene and pathway level responses.
The study identified significant upregulation of pathways related to synaptic function and lipid metabolism following lead and fluoxetine exposure, respectively. Additionally, fluoxetine exposure elevated several fatty acids, which was validated using mass spectrometry-based metabolomics. The study demonstrated that FFED can be used for multiplexed transcriptomic analyses to detect relevant pathway-level changes in human neural development caused by low-grade environmental risk factors.
The findings of this study have significant implications for understanding the mechanisms of neurodevelopmental disorders, such as autism spectrum disorder (ASD), and the role of environmental exposures in their etiology. The study showed that RNA sequencing, coupled with FFED, is an efficient and multiplexable method for studying the effects of environmental factors and their mixtures on molecular outcomes. These findings will help to develop efficient methods to estimate the effects of environmental factors during early human development.
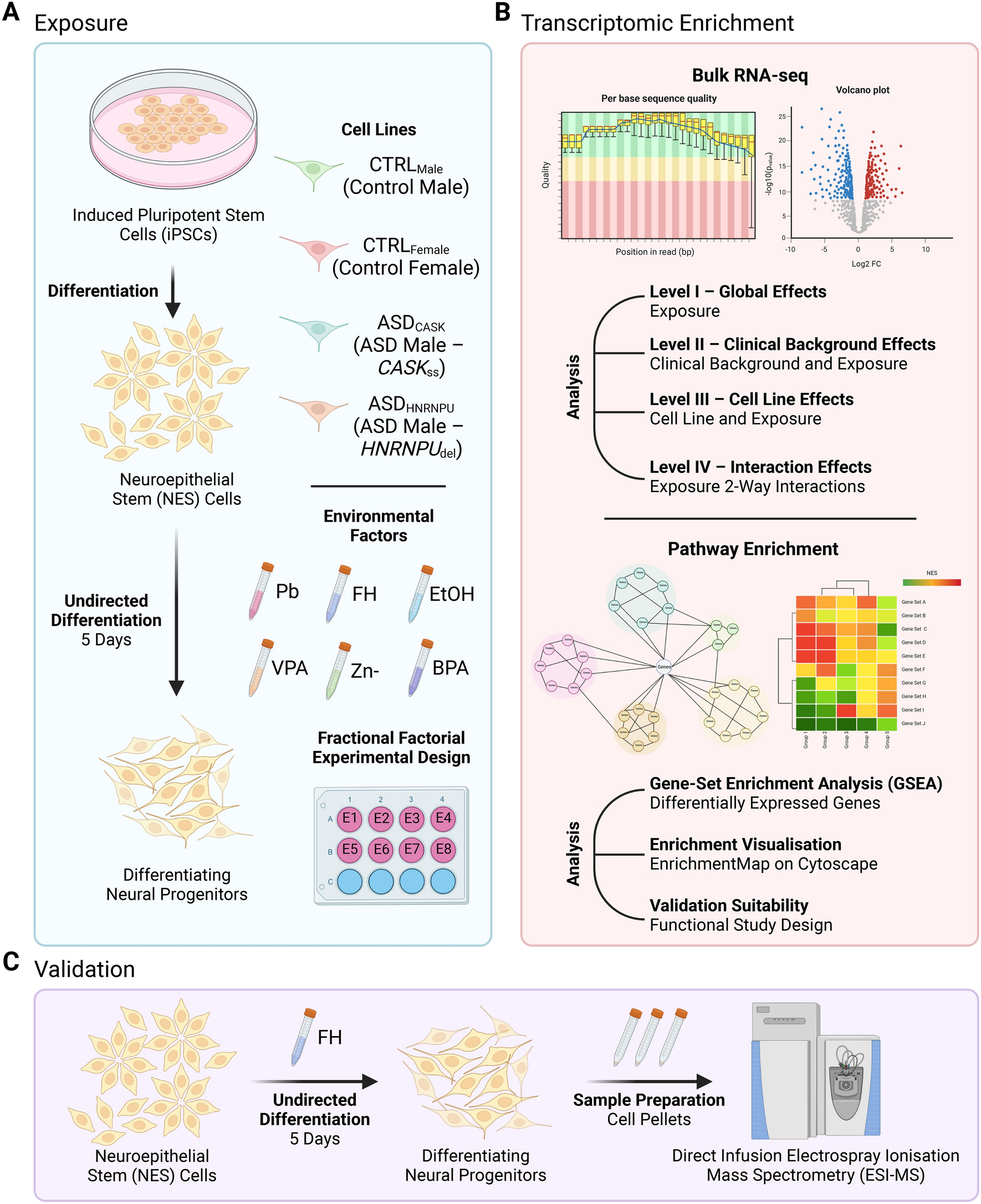
Overview of study plan using fractional factorial experimental design (FFED)
(A) Exposure of neuroepithelial stem (NES) cells derived from human induced pluripotent stem cells (iPSCs) during neural progenitor differentiation for 5 days. Four cell lines, neurotypical controls (CTRLMale, CTRLFemale) and males with autism spectrum disorder (ASD) diagnoses (ASDCASK: CASK splice site variant, ASDHNRNPU: HNRNPU deletion), were exposed to six environmental factors during differentiation, namely lead (Pb), fluoxetine hydrochloride (FH), ethanol (EtOH), valproic acid (VPA), bisphenol A (BPA) and zinc deficiency (Zn-). (B) Using RNA samples from (A), bulk RNA-sequencing was performed followed by differential gene expression analysis for global effects, clinical background effects, cell line effects and interaction effects. Using this, pathway enrichment and network visualisation was done. (C) Enriched pathways identified from (B) following FH exposure were detected using direct infusion electrospray ionisation mass spectrometry (ESI–MS) and quantified across the different levels of analyses. (Created with BioRender.com).
Arora A, Becker M, Marques C et al. (2023) Screening autism-associated environmental factors in differentiating human neural progenitors with fractional factorial design-based transcriptomics. Sci Rep 13, 10519. [article]