Our understanding of nerve regeneration can be enhanced by delineating its underlying molecular activities at single-neuron resolution in model organisms such as Caenorhabditis elegans. Existing cell isolation techniques cannot isolate neurons with specific regeneration phenotypes from C. elegans. Researchers from The University of Texas at Austin have developed femtosecond laser microdissection (fs-LM), a single-cell isolation method that dissects specific cells directly from living tissue by leveraging the micrometer-scale precision of fs-laser ablation. The researchers show that fs-LM facilitates sensitive and specific gene expression profiling by single-cell RNA sequencing (scRNA-seq), while mitigating the stress-related transcriptional artifacts induced by tissue dissociation. scRNA-seq of fs-LM isolated regenerating neurons revealed transcriptional programs that are correlated with either successful or failed regeneration in wild-type and dlk-1 (0) animals, respectively. This method also allowed studying heterogeneity displayed by the same type of neuron and found gene modules with expression patterns correlated with axon regrowth rate. These results establish fs-LM as a spatially resolved single-cell isolation method for phenotype-to-genotype mapping.
fs-LM isolates intact neuronal somas with reduced stress-related
transcriptional artifacts for scRNA-seq
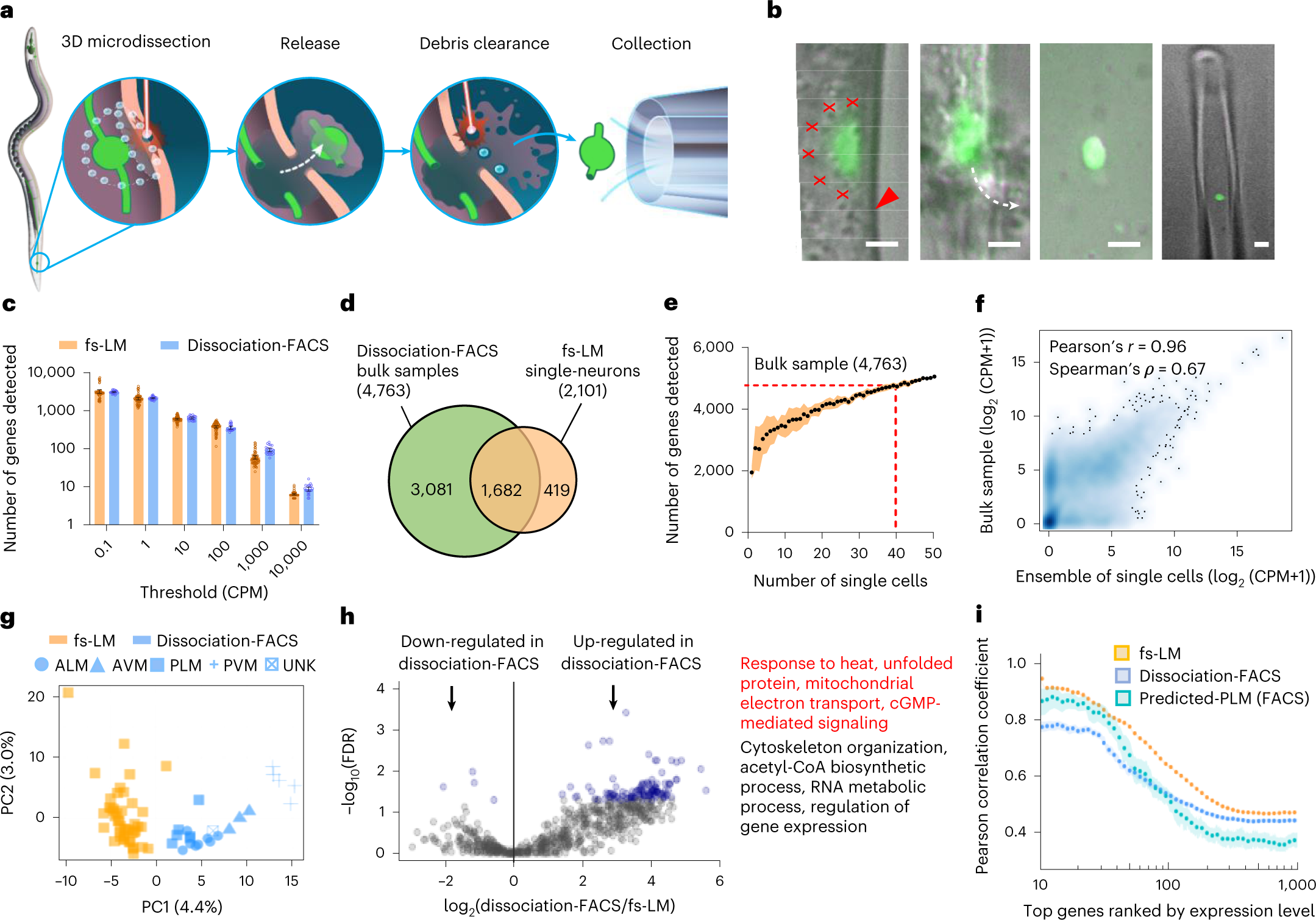
a, Schematic depiction of the fs-LM method. b, Microscope images during fs-LM of a GFP-labeled PLM neuron of C. elegans in the order shown in a. Red crosses show locations of laser ablation spots. Red arrowhead shows the location of cuticle ablation. White dashed line shows the trajectory of neuron release (n = 123 of 384 attempts). Scale bars, 2 µm. c, Number of genes detected at various thresholds in single PLM neurons (n = 64 uninjured neurons of wild-type and dlk-1(0) animals) isolated by fs-LM and single TRNs (n = 23) isolated by the dissociation-FACS method. Data are presented as mean ± 95% CI (confidence interval) d, Overlap between genes identified in fs-LM isolated single PLM neurons and three bulk samples (>5,000 dissociation-FACS-isolated TRNs from wild-type animals) at CPM ≥ 1. e, Number of unique genes detected as the number of fs-LM isolated neurons increases. Data are presented as mean ± s.d. f, Correlation of gene expression profiles between the ensemble of 40 fs-LM isolated neurons and bulk sample. g, Principal component (PC) analysis on gene expression profiles in single neurons isolated from wild-type animals by fs-LM (n = 45) and dissociation-FACS (n = 23) methods. The dissociation-FACS-isolated neurons were labeled as ALM, AVM, PLM and PVM, as identified using the neuron-specific CeNGEN data. UNK represents an unknown neuron. h, Volcano plot showing DEGs with blue highlighting the substantially down- and up-regulated genes (FDR < 0.05) in dissociation-FACS-isolated neurons. The list shows representative GO terms associated with the up-regulated genes, including key functions related to stress response (highlighted in red). i, Correlation among gene expression profiles of single neurons isolated by fs-LM or dissociation-FACS methods or predicted-PLM neurons isolated by the dissociation-FACS method, showing a statistically higher correlation among fs-LM isolated neurons (two-sided Wilcoxon rank sum test, P = 2.87 × 10−5 between fs-LM and predicted-PLM neurons for the top ten genes). Data are presented as mean ± 95% CI.
Zhao P, Mondal S, Martin C et al. (2023) Femtosecond laser microdissection for isolation of regenerating C. elegans neurons for single-cell RNA sequencing. Nat Methods [Epub ahead of print]. [abstract]