Multiple myeloma (MM) is a complex and diverse form of cancer that affects plasma cells, a type of white blood cell crucial for the immune system. Despite advancements in medical research, accurately predicting the outcomes and progression of MM remains challenging. Traditional prognostic models have limitations in capturing the full spectrum of variability in disease outcomes. However, a recent breakthrough leveraging single-cell RNA-sequencing data has led to the development of a novel plasma cell gene signature, offering new hope for improved prognostic assessment in MM.
In this groundbreaking study, researchers from Baylor College of Medicine analyzed single-cell RNA-sequencing data to identify a unique gene signature specific to plasma cell malignancy (PBM). The resulting PBM score was then evaluated and validated using data from five independent myeloma studies, encompassing a total of 2115 samples, including MM patients, individuals with precursor conditions like monoclonal gammopathy of undetermined significance (MGUS) and smoldering MM, as well as healthy controls.
Overview of the study
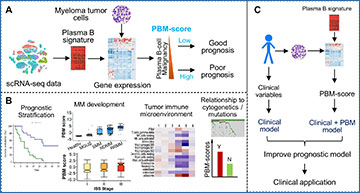
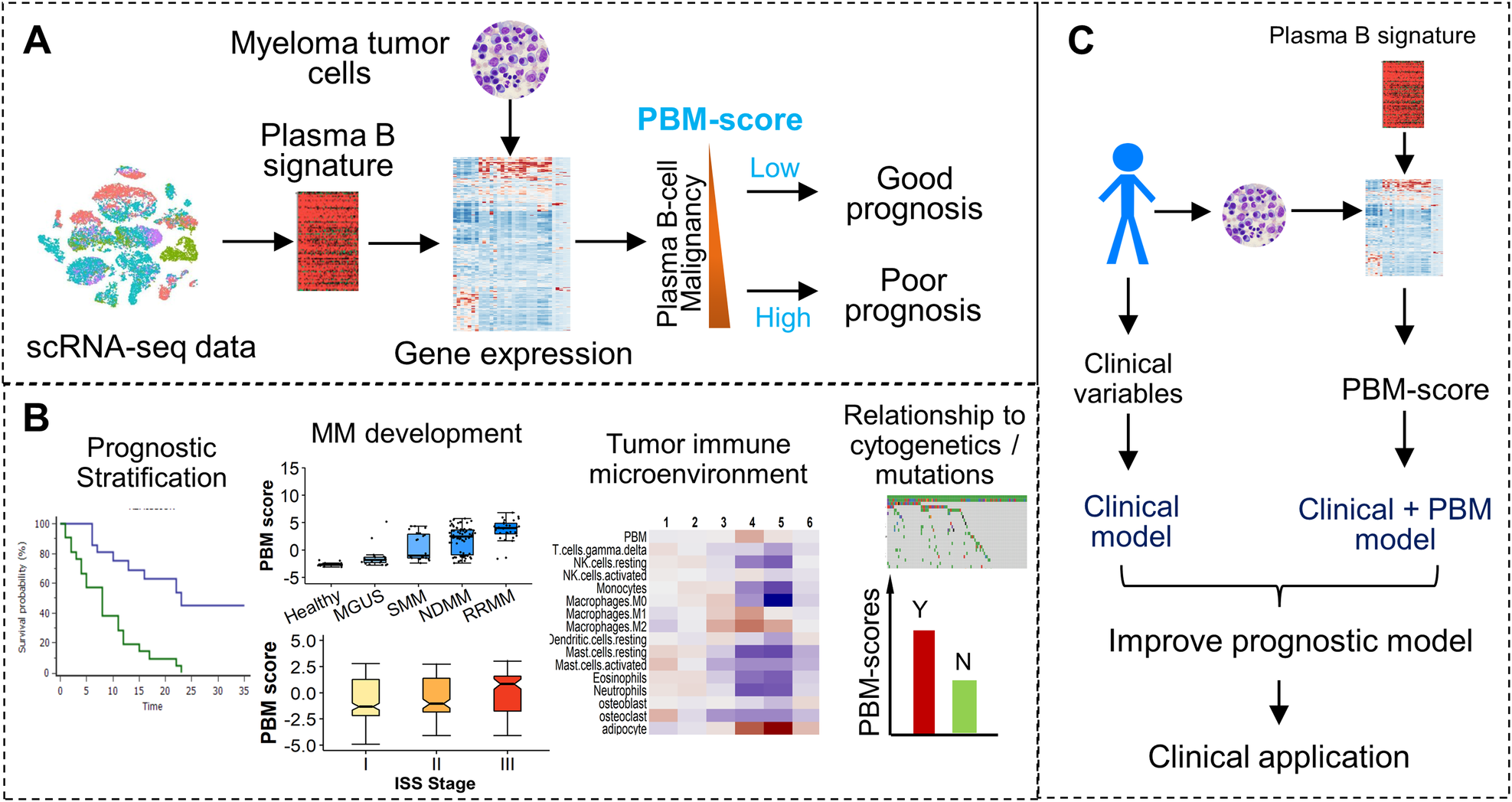
A A plasma B-cell signature was defined to quantify the magnitude of malignancy of MM samples. The resulting plasma B malignancy (PBM) score is associated with the patient’s prognosis. B The PBM scores of CD138+ cells were found to be used for prognosis prediction, characterize the MM development and progression, and be correlated with the patient’s immune microenvironment and cytogenetic abnormalities. C The PB signature was incorporated into the prognostic model to improve prediction accuracy for better clinical application.
The study revealed compelling associations between the PBM score and various aspects of MM, including disease stage, progression, and clinical outcomes. Specifically, higher PBM scores were significantly correlated with more advanced stages of plasma cell dyscrasias, indicating a potential role in disease severity assessment. Moreover, patients with MM exhibiting higher PBM scores had shorter overall survival rates, highlighting the prognostic value of this novel gene signature.
Importantly, the prognostic impact of the PBM score remained significant even after accounting for established staging systems like the International Staging System (ISS) and Revised ISS (R-ISS), suggesting its independent predictive power.
Furthermore, downstream analysis revealed intriguing associations between elevated PBM scores and other MM-related factors, including cytogenetic abnormalities, TP53 mutations, and alterations in the tumor immune microenvironment. These insights offer valuable clues for understanding the underlying mechanisms of MM progression and immune response modulation.
The development of the PBM score represents a significant advancement in the field of MM research. By providing a refined and independent prognostic indicator, this novel gene signature holds promise for improving risk stratification and guiding treatment decisions for MM patients. Clinically, the integration of the PBM score into existing diagnostic and therapeutic protocols may pave the way for more personalized and effective management strategies in MM.
The identification and validation of the PBM score mark a pivotal milestone in our quest to better understand and manage multiple myeloma. This innovative approach not only enhances our ability to predict disease outcomes but also offers insights into the molecular mechanisms driving MM progression. Moving forward, continued research and clinical validation of the PBM score may ultimately lead to improved patient outcomes and better-targeted therapies for this complex hematological malignancy.
Li JR, Arsang-Jang S, Cheng Y, Sun F, D’Souza A, Dhakal B, Hari P, Huang Q, Auer P, Li Y, Urrutia R, Zhan F, Shaughnessy JD Jr, Janz S, Dong J, Cheng C. (2024) Enhancing prognostic power in multiple myeloma using a plasma cell signature derived from single-cell RNA sequencing. Blood Cancer J 14(1):38. [article]