Double-stranded RNAs (dsRNAs) are potent triggers of innate immune responses upon recognition by cytosolic dsRNA sensor proteins. Identification of endogenous dsRNAs helps to better understand the dsRNAome and its relevance to innate immunity related to human diseases.
UCLA researchers have developed dsRID (double-stranded RNA identifier), a machine-learning-based method to predict dsRNA regions in silico, leveraging the power of long-read RNA-sequencing (RNA-seq) and molecular traits of dsRNAs. Using models trained with PacBio long-read RNA-seq data derived from Alzheimer’s disease (AD) brain, the researchers show that their approach is highly accurate in predicting dsRNA regions in multiple datasets. Applied to an AD cohort sequenced by the ENCODE consortium, they characterize the global dsRNA profile with potentially distinct expression patterns between AD and controls. Together, they show that dsRID provides an effective approach to capture global dsRNA profiles using long-read RNA-seq data.
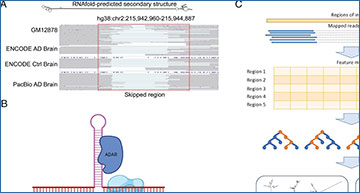
Overview of the dsRID method
(A) An example region showing internal skipping that occurs in multiple datasets. Top: RNAfold-predicted structure of the genomic region. Bottom: IGV plots of mapped reads from four datasets. Ctrl: control. (B) Schematic diagram showing the hypothesis of template skipping due to double-stranded structure and ADAR binding (created by Biorender). (C) Schematic diagram for the steps in dsRID
Availability – Software implementation of dsRID, and genomic coordinates of regions predicted by dsRID in all samples are available at the GitHub repository: https://github.com/gxiaolab/dsRID.
Yamamoto R, Liu Z, Choudhury M, Xiao X. (2023) dsRID: in silico identification of dsRNA regions using long-read RNA-seq data. Bioinformatics 39(11):btad649. [article]