Owing to the nondeterministic and nonlinear nature of gene expression, the steady-state intracellular protein abundance of a clonal population forms a distribution. The characteristics of this distribution, including expression strength and noise, are closely related to cellular behavior. However, quantitative description of these characteristics has so far relied on arrayed methods, which are time-consuming and labor-intensive.
To address this issue, Tsinghua University researchers have developed a deep-learning-assisted Sort-Seq approach (dSort-Seq), enabling high-throughput profiling of expression properties with high precision. They demonstrated the validity of dSort-Seq for large-scale assaying of the dose-response relationships of biosensors. In addition, the researchers comprehensively investigated the contribution of transcription and translation to noise production in Escherichia coli, from which they found that the expression noise is strongly coupled with the mean expression level. The researchers also found that the transcriptional interference caused by overlapping RpoD-binding sites contributes to noise production, which suggested the existence of a simple and feasible noise control strategy in E. coli.
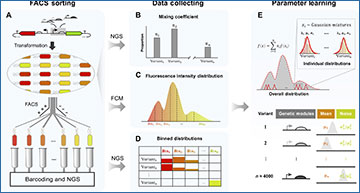
Schematic overview of the dSort-Seq data workflow
(A) During Sort-Seq, a library with different expression patterns is sorted into customized bins based on the fluorescence intensity value. (B) The mixing coefficients are quantified via NGS. (C) The overall fluorescence density is measured by FCM, and the sorting boundaries are specified on the basis of the overall fluorescence intensity density. (D) The read count number across all bins as quantified by NGS reveals the binned distribution of each variant in the library. (E) Through parameter learning, the mean, expression noise, and their relationships can be precisely identified. μ, mean; σ, SD.
Availability – https://github.com/fenghuibao/dSort-Seq
Feng H, Li F, Wang T, Xing XH, Zeng AP, Zhang C. (2023) Deep-learning-assisted Sort-Seq enables high-throughput profiling of gene expression characteristics with high precision. Sci Adv 9(45):eadg5296. [article]