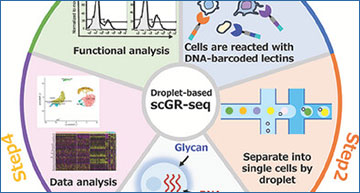
Plate-based single-cell glycan and RNA sequencing (scGR-seq) is previously developed to realize the integrated analysis of glycome and transcriptome in single cells. However, the sample size is limited to only a few hundred cells. Here, a droplet-based scGR-seq is developed to address this issue by adopting a 10x Chromium platform to simultaneously profile ten thousand cells’ glycome and transcriptome in single cells. To establish droplet-based scGR-seq, researchers at the National Institute of Advanced Industrial Science and Technology, Japan have performed a comparative analysis of two distinct cell lines: pancreatic ductal adenocarcinoma cells and normal pancreatic duct cells. Droplet-based scGR-seq revealed distinct glycan profiles between the two cell lines that showed a strong correlation with the results obtained by flow cytometry. Next, droplet-based scGR-seq is applied to a more complex sample: peripheral blood mononuclear cells (PBMC) containing various immune cells. The method can systematically map the glycan signature for each immune cell in PBMC as well as glycan alterations by cell lineage. Prediction of the association between the glycan expression and the gene expression using regression analysis ultimately leads to the identification of a glycan epitope that impacts cellular functions. In conclusion, the droplet-based scGR-seq realizes the high-throughput profiling of the distinct cellular glyco-states in single cells.
Keisham S, Saito S, Kowashi S, Tateno H. (2024) Droplet-Based Glycan and RNA Sequencing for Profiling the Distinct Cellular Glyco-States in Single Cells. Small Methods [Epub ahead of print]. [abstract]