Understanding the intricate workings of the kidney and how it responds to injury is crucial for developing effective treatments for kidney-related diseases. A recent study harnesses the power of emerging spatially resolved transcriptomics technologies to delve deep into the molecular landscape of healthy and injured mouse kidneys.
Using a cutting-edge method called direct RNA hybridization-based in situ sequencing (dRNA HybISS), researchers from the Washington University in St. Louis School of Medicine compared gene expression patterns in male and female healthy mouse kidneys, as well as in male kidneys undergoing injury and repair. By analyzing a panel of 200 genes, they uncovered dynamic alterations in cell states during injury and repair processes.
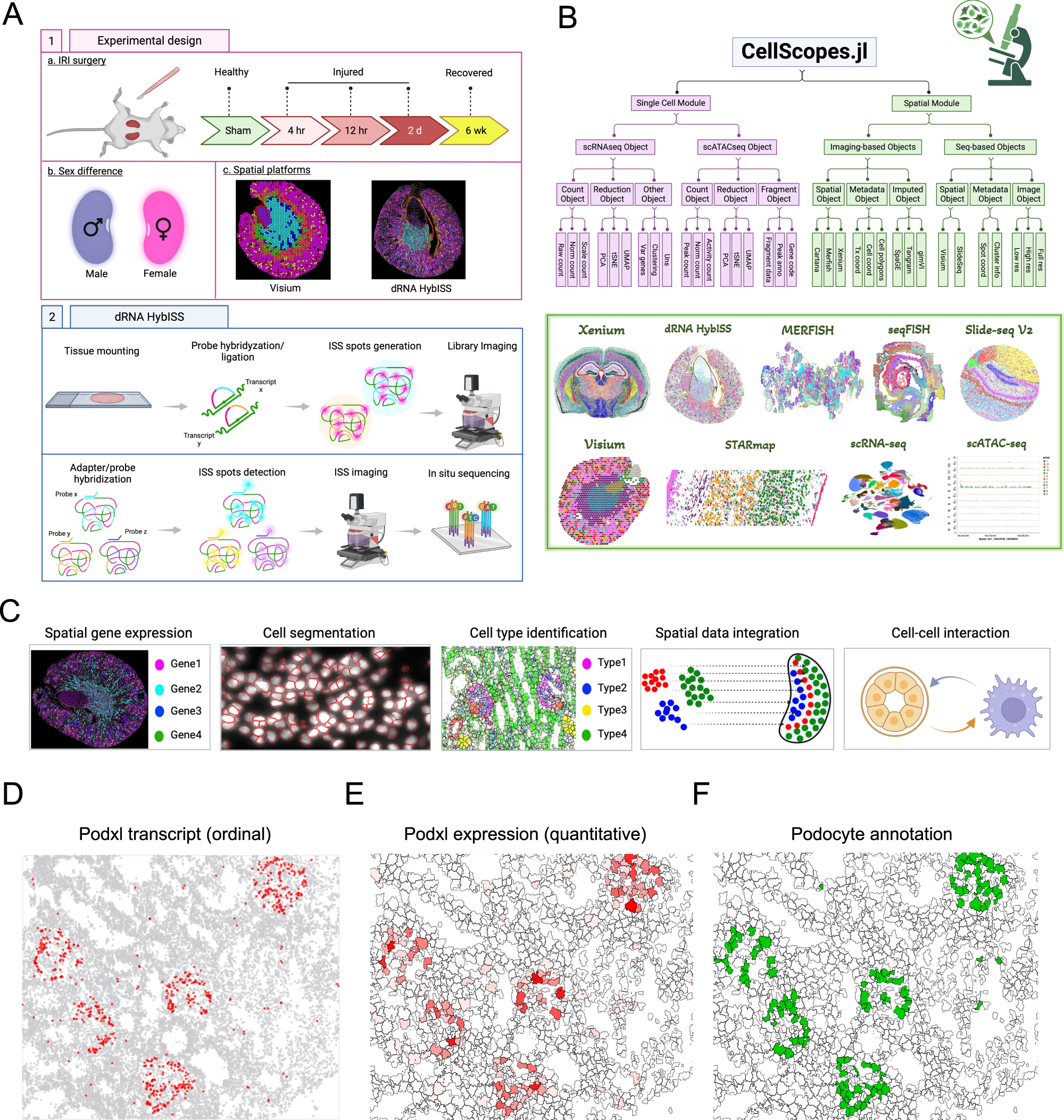
To analyze this wealth of data, the researchers developed a new computational pipeline called CellScopes. This innovative tool enables rapid analysis, integration of multiple omics datasets, and visualization of spatially resolved transcriptomic data. Through CellScopes, researchers were able to identify 13 distinct kidney cell types within specific kidney niches and track changes in cell states over the course of injury and repair.
dRNA HybISS workflow, experimental design and computational analysis
A Schematic of using dRNA HybISS to study AKI. Schematic was created with BioRender. B A Julia package, CellScopes.jl, was developed for spatial data processing, analysis and visualization. Image was created with BioRender. C Outline of the spatial analysis pipeline used in this study, created with BioRender. D–F CellScopes.jl allows for visualization of gene expression on cells as data points (D), segmented polygons (E), and cell-type annotations (F).
One intriguing finding of the study was the identification of cell-cell interactions between leukocytes and kidney parenchyma. At late stages after injury, certain immune cells known as C3+ leukocytes were found to be enriched near pro-inflammatory proximal tubule cells that had failed to repair properly.
Moreover, by integrating single-nucleus RNA sequencing (snRNA-seq) data from the same injury and repair samples, researchers were able to impute the spatial localization of genes not directly measured by dRNA HybISS. This integration of datasets provided a more comprehensive understanding of the molecular events underlying kidney injury and repair processes.
Overall, this study highlights the power of spatially resolved transcriptomics in unraveling the complex dynamics of kidney health and disease. By shedding light on the molecular interactions within the kidney microenvironment, researchers move closer to developing targeted therapies for kidney-related disorders, ultimately improving patient outcomes and quality of life.
Availability – A Julia package, CellScopes.jl, for analyzing various spatial transcriptomics data were available at https://github.com/TheHumphreysLab/CellScopes.jl.
Wu H, Dixon EE, Xuanyuan Q et al.(2024) High resolution spatial profiling of kidney injury and repair using RNA hybridization-based in situ sequencing. Nat Commun 15, 1396. [article]